10x Genomics
Chromium Single Cell Gene Expression
Cell Ranger6.4 (latest), printed on 07/12/2025
Loupe Browser Tutorial
This set of tutorials review the major analysis capabilities Loupe Browser provides for analyzing 3' Single Cell Gene Expression, Feature Barcode data, and 5' V(D)J and Single Cell Gene Expression data.
- Navigation
- Significant Features
- Identifying Cell Types
- Exploring Substructure
- Identifying Cell Subtypes
- Filtering & Reclustering
- Sharing Results
- Interoperability
- V(D)J Clonotype Analysis
- Feature Barcode Data
- Capturing Neutrophils
- Analyzing Public Data
Installing Loupe Browser
To use Loupe Browser, follow the directions on the Downloads page to download and install the software on either macOS or Windows.
Loupe Browser tutorials
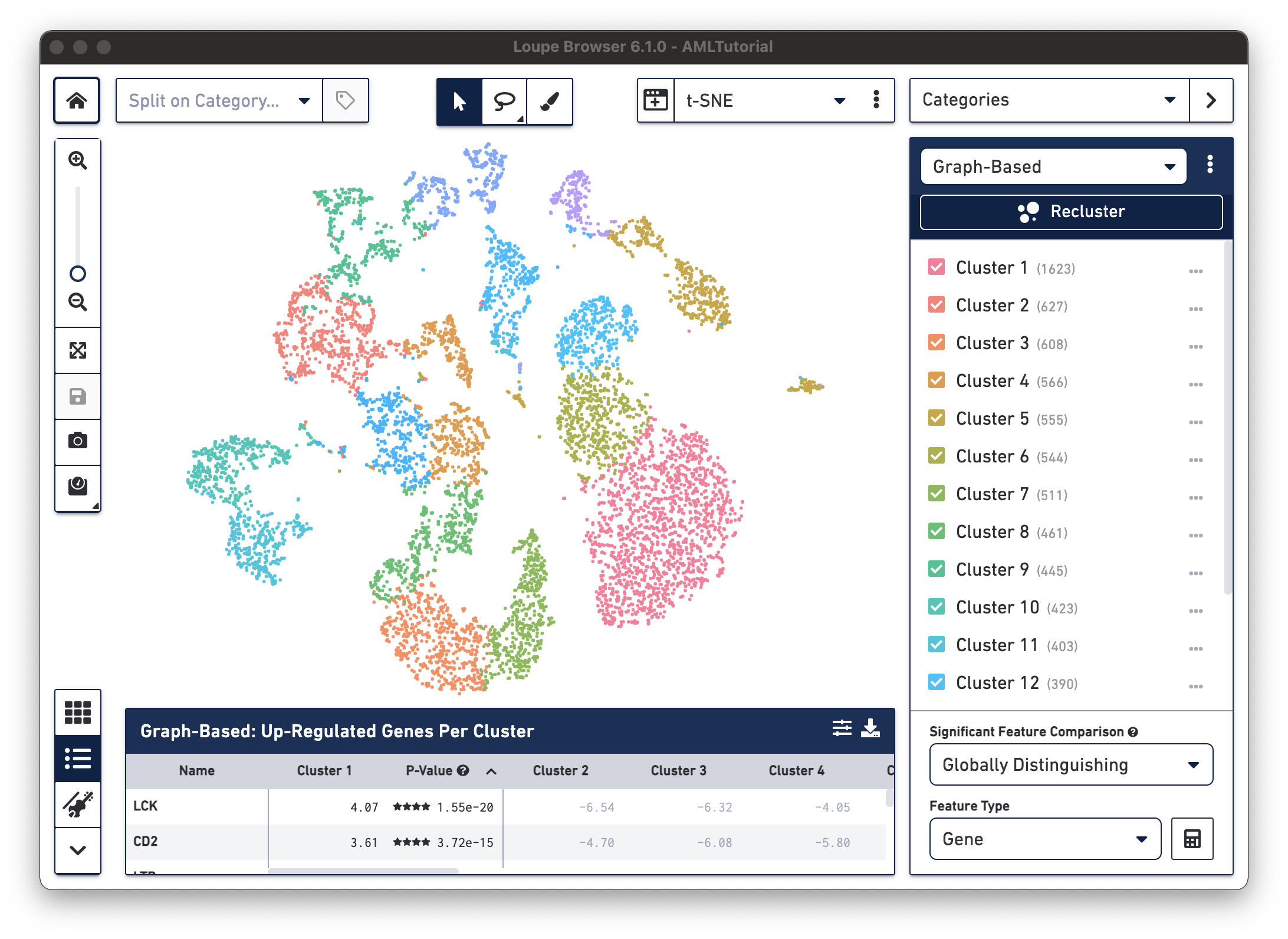
Open Loupe Browser by double-clicking on the application icon. Then click on the
AMLTutorial file from the list of Recent Files. You should see a screen with the t-SNE projection for the cells from
the AML dataset.
Once you have installed Loupe Browser, you can follow the tutorials using the AML example dataset:
- Learn about its capabilities in the navigation tutorial.
- Learn how to analyze output data from Cell Ranger, starting with finding significant features.
Dataset description
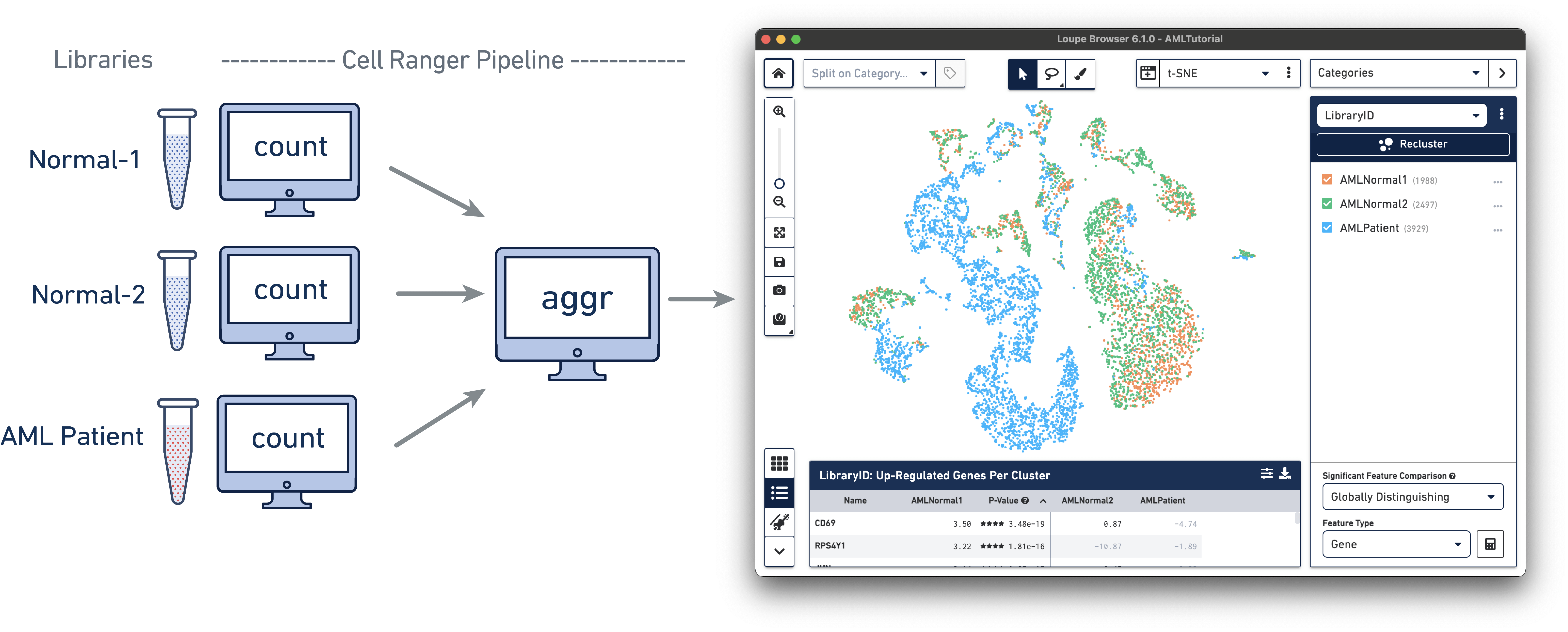
The AML Tutorial dataset contains the results of a Cell Ranger aggr pipeline analysis run for three samples, including two healthy control samples of frozen human bone marrow mononuclear cells and a pre-transplant sample from a patient with acute myeloid leukemia (AML). This dataset was generated in collaboration with the Fred Hutchinson Cancer Research Center, and referenced in the Nature Communications publication, "Zheng et al, Massively parallel digital transcriptional profiling of single cells" (2017; doi:10.1038/ncomms14049).
Sharing usage data
Starting with Loupe Browser v6.1, users may choose to share anonymized non-biological usage data (i.e. no identifiable sample information such as files, cluster names, or any open text fields are tracked) to improve Loupe Browser performance. Sharing of usage data is optional, and users can change whether they want to share usage data any time. The user preference for usage data sharing will carry over within a major version (e.g. all 6.Y versions) but the user will be asked to confirm their preferences for each major version update.
In case you wish to change the usage data sharing setting, click Manage Usage Data from the Help menu and change the preference in the pop-up window.