10x Genomics
Single Cell Gene Expression
Cell Ranger, printed on 07/16/2025
System Requirements
Table of Contents
Hardware
Cell Ranger pipelines run on Linux systems that meet these minimum requirements:
- 8-core Intel or AMD processor (16 cores recommended)
- 64GB RAM (128GB recommended)
- 1TB free disk space
- 64-bit CentOS/RedHat 7.0 or Ubuntu 14.04 (deprecated November, 2021); See the 10x Genomics OS Support page for details.
Note: Cell Ranger v6.1 was the last version that supported CentOS/RedHat 6 or Ubuntu 12.04.
| The minimum requirement of 64GB RAM will allow cellranger aggr to aggregate up to 250k cells, more memory will be required beyond that. |
In order to run in cluster mode, the cluster needs to meet these additional minimum requirements:
- Shared filesystem (e.g. NFS)
- SGE or LSF batch scheduling system
Software dependencies
In order to run cellranger mkfastq, the following software needs to be installed:
- Illumina® bcl2fastq: bcl2fastq must be version 2.20 or higher.
Resource limits
- Cell Ranger runs with
--jobmode=localby default, using 90% of available memory and all available cores. To restrict resource usage, please see the--localmemand--localcoresflags for cellranger count at the link here for more information. - Many Linux systems have default user limits (ulimits) for maximum open files and maximum user processes as low as 1024 or 4096. Because Cell Ranger spawns multiple processes per core, jobs that use a large number of cores can exceed these limits. 10x Genomics recommends higher limits.
| Limit | Recommendation |
|---|---|
| user open files | 16k |
| system max files | 10k per GB RAM available to Cell Ranger |
| user processes | 64 per core available to Cell Ranger |
How memory affects runtime
The following plots are based on time trials using Amazon EC2 instances and version 7.0 of Cell Ranger. Observed performance will be impacted by other factors besides memory, including library type (whole transcriptome vs. targeted), read depth, number of cells, number of cores/threads, and I/O. The plots below are meant to convey how increasing memory beyond the minimum and recommended levels may impact runtime for large datasets run with cellranger multi, cellranger count, and cellranger aggr, but may not be representative of your dataset.
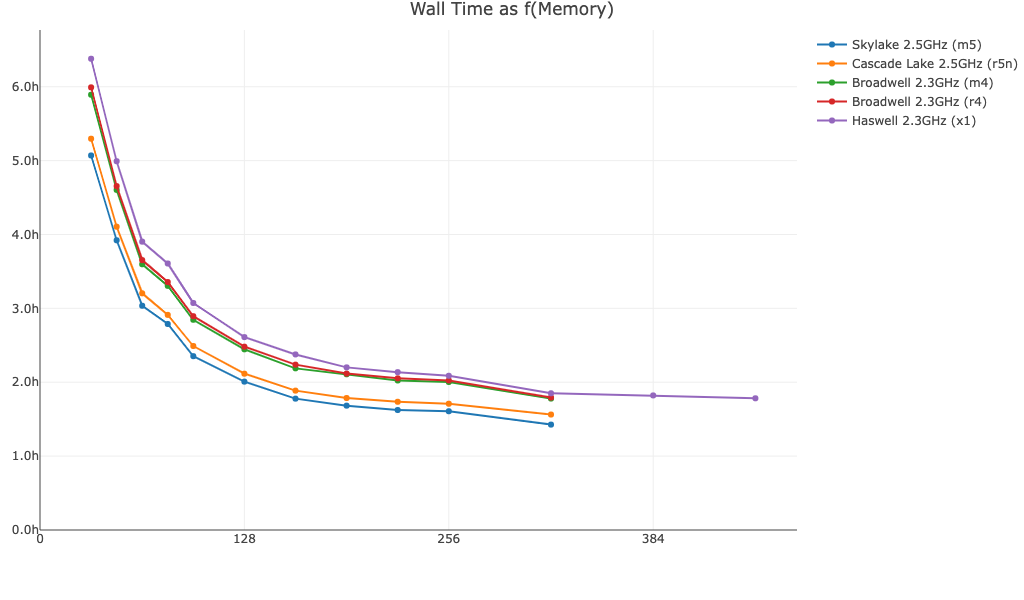
Shown below is cellranger multi wall time as a function of memory for a 60k cell 3' Cell Multiplexing dataset:
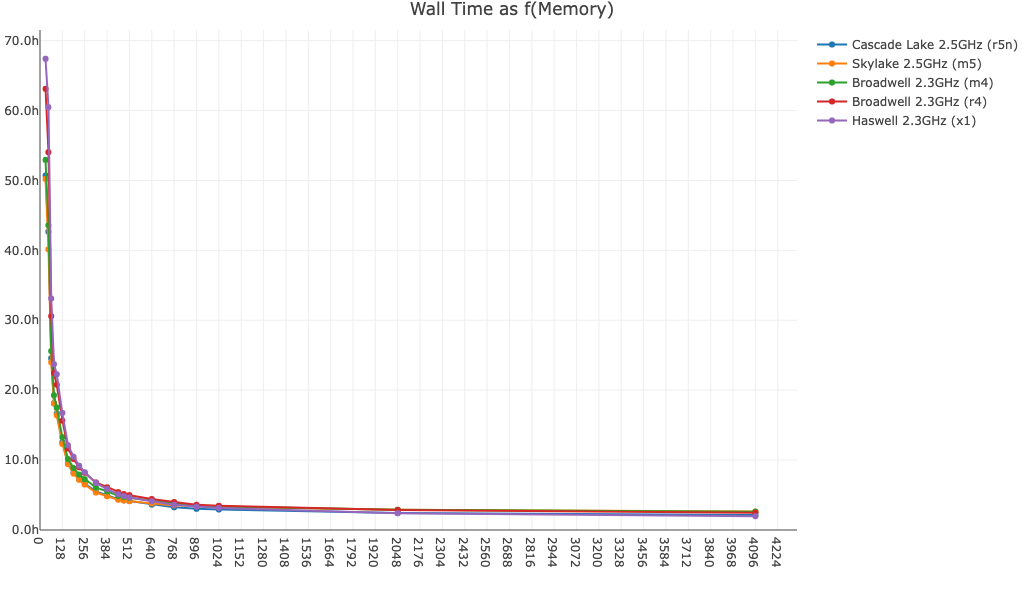
Here is cellranger multi wall time as a function of memory for a 16-plex Fixed RNA Profiling dataset run with BAM file generation on and off:
- BAM file generated :
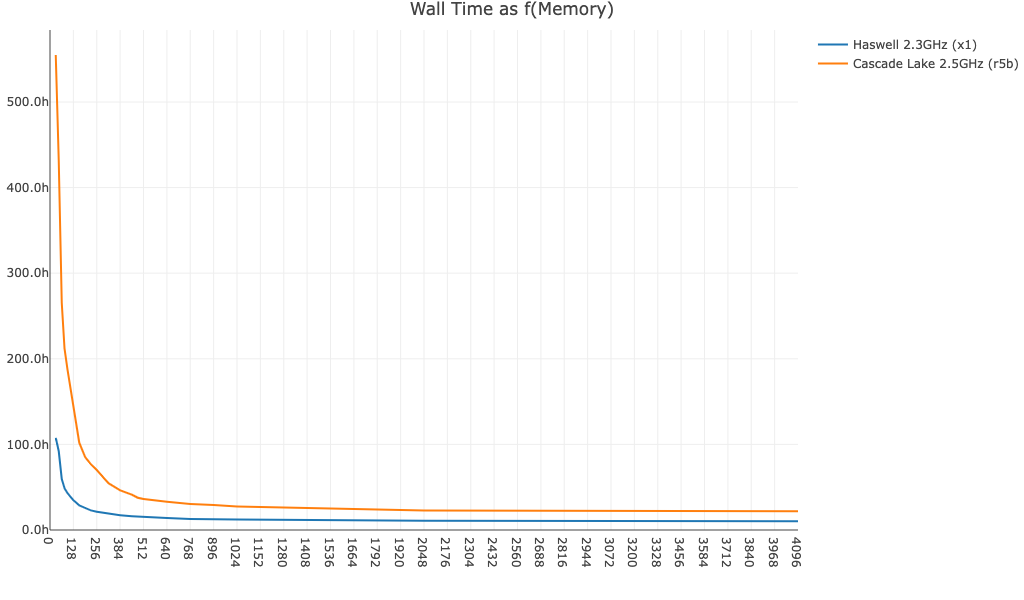
- No BAM file generated (
no-bam,true):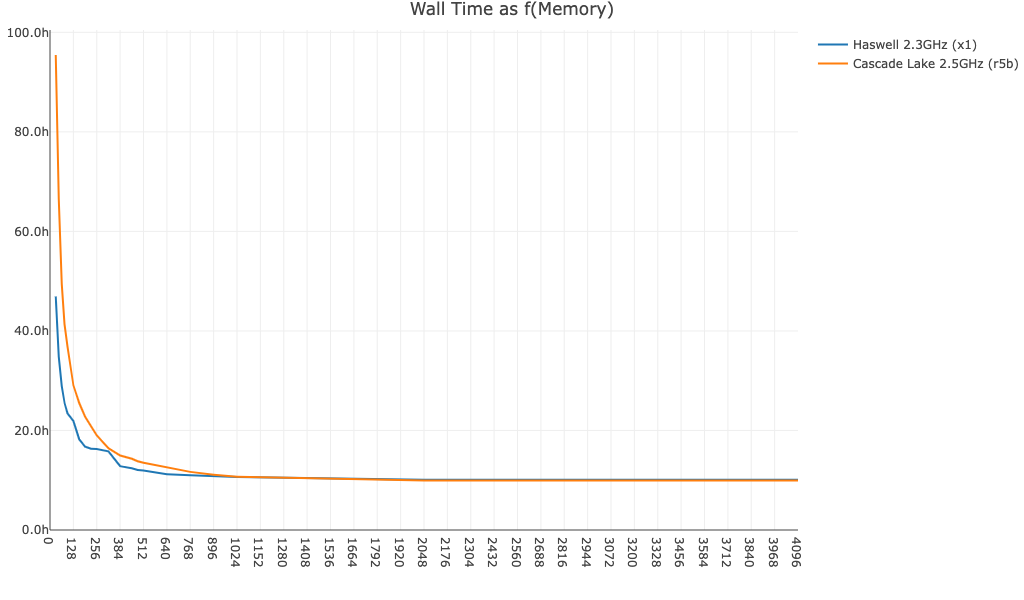
Here is cellranger count wall time as a function of memory for a 20k PBMC high-throughput dataset:
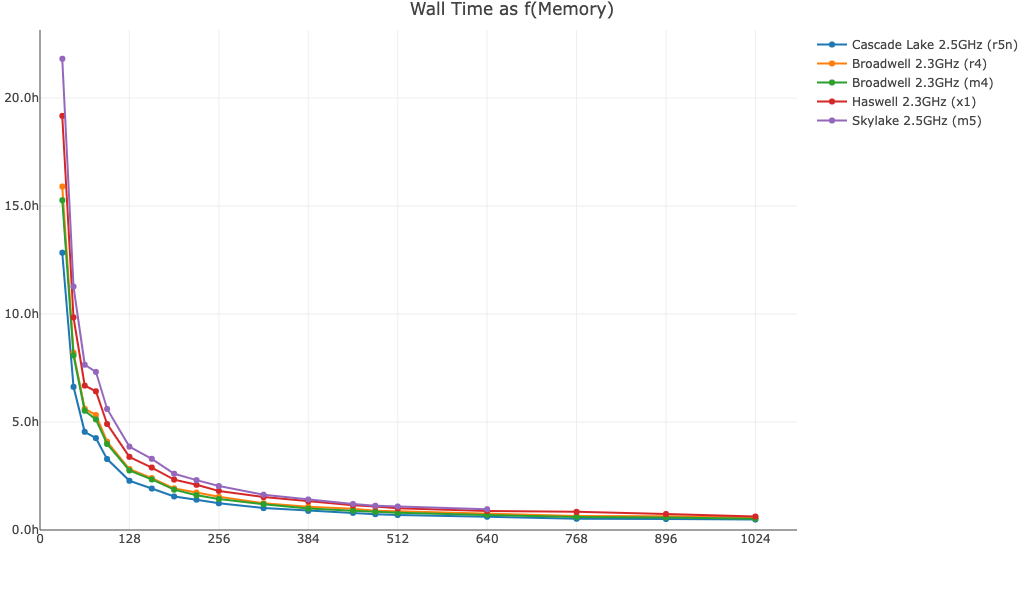
Here is cellranger aggr wall time as a function of available memory for a 250k cell dataset. In general, performance can be improved by allocating more than the minimum 64GB of memory to the pipeline.