10x Genomics
Chromium Single Cell Immune Profiling
Cell Ranger3.0, printed on 07/12/2025
Single Cell V(D)J + 5′ Gene Expression + Feature Barcoding for Cell Surface Protein Analysis
Using the Chromium Single Cell Immune Profiling Solution with Feature Barcoding technology, simultaneously measure V(D)J, gene expression, and cell surface protein expression in the same cells. This allows users to analyze the adaptive immune response inside complex tissue samples, observe the effects of immunotherapies, and investigate complex autoimmune, infectious, and other immune diseases.
See V(D)J and Feature Barcoding technology for an overview of Single Cell Immune Profiling with Feature Barcoding technology.
Workflow
Download the latest versions of Cell Ranger, Loupe Cell Browser, and Loupe V(D)J Browser to get started. The workflow for generating V(D)J, Gene Expression and Cell Surface Protein libraries from 10x Barcoded, amplified DNA molecules is illustrated below.
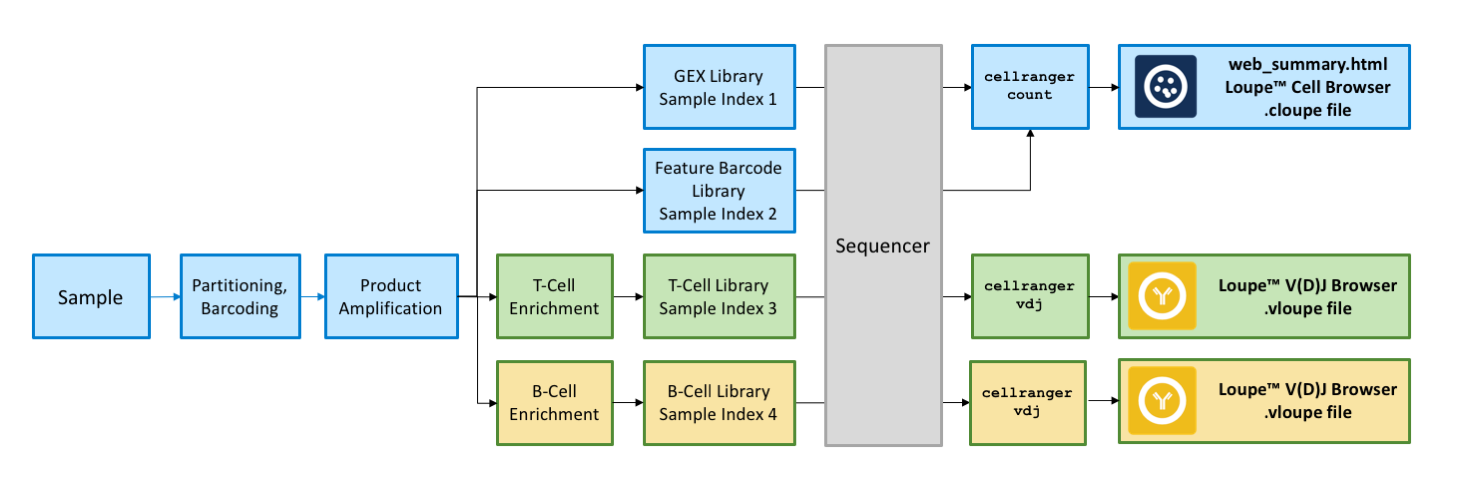
Note that the sample indices used for V(D)J Enriched and 5’ Gene Expression libraries are unique from the sample indices used for Cell Surface Protein libraries. For V(D)J Enriched and 5’ Gene Expression libraries, use ONLY Chromium i7 Sample Index Plate. For Cell Surface Protein library, use ONLY Chromium i7 Sample Index Plate N, Set A. Consider sample index compatibility when pooling different libraries; unique sample index for each of the pooled libraries is required.
For the complete workflow, please consult the Chromium™ Single Cell V(D)J Reagent Kits User Guide and the Chromium™ Single Cell V(D)J Reagent Kits with Feature Barcoding technology for Cell Surface Protein
Sequencing
Up to four different library types with unique sample indices can be generated for sequencing:
- V(D)J Enriched library (TCR and/or Ig)
- 5’ Gene Expression library
- Cell Surface Protein library
All these libraries may be pooled for sequencing, taking into account the differences in depth requirements between the pooled libraries. Here are the recommended sequecing configuration and read depth for each library type. When pooling multiple library types, use the longest read length configuration among the types being pooled.
| Library Type | R1 Cycles | I7 Cycles | R2 Cycles | Minimum Reads per Cell |
|---|---|---|---|---|
| V(D)J T/B cell Enriched | 150bp | 8bp | 150bp | 5,000 |
| 5′ Gene Expression | 26bp | 8bp | 91bp | 20,000 |
| Feature Barcode Library | 26bp | 8bp | 25bp | 5,000 |
The compatibility of the listed sequencers has been verified by 10x Genomics. Some variation in assay performance is expected based on sequencer choice.
- MiSeq
- NextSeq 500/550*
- HiSeq 2500 (Rapid Run)
- HiSeq 3000/4000
- NovaSeq
For additional information, consult the Single Cell Immune Profiling Sequencing page.
Running Cell Ranger
Assume a sample sheet was used that yielded FASTQs with sample name prefixes SampleGEX, SampleT and SampleB. To process the three different library types, run separate instances of the new Cell Ranger 3.0 pipelines:
cellranger vdjto analyze data from the SampleT and SampleB librariescellranger countto analyze gene expression from the SampleGEX library.cellranger countperforms Feature Barcoding analysis simultaneously with Gene Expression analysis. Consult Cell Ranger Feature Barcoding Analysis in the Gene Expression section for more details.
To analyze the data from the enriched T cell or B cell libraries, run the cellranger vdj pipeline:
$ cd /home/jdoe/runs $ cellranger vdj --id=GEX_VDJ_SampleT \ --reference=/opt/refdata-cellranger-vdj-GRCh38-alts-ensembl-2.0.0 \ --fastqs=/home/jdoe/mkfastq_pipeline/outs/fastq_path \ --sample=SampleT \
$ cd /home/jdoe/runs $ cellranger vdj --id=GEX_VDJ_SampleB \ --reference=/opt/refdata-cellranger-vdj-GRCh38-alts-ensembl-2.0.0 \ --fastqs=/home/jdoe/mkfastq_pipeline/outs/fastq_path \ --sample=SampleB \
These pipelines will generate a Loupe V(D)J Browser .vloupe file, in addition to a variety of files
to describe V(D)J clonotypes, consensus sequences, and contigs. For more details on running
the V(D)J analysis pipeline, consult the Cell Ranger V(D)J documentation.
To analyze the data from the gene expression library, run the cellranger count pipeline:
$ cd /home/jdoe/runs $ cellranger count --id=GEX_VDJ_SampleGEX \ --transcriptome=/opt/refdata-cellranger-GRCh38-1.2.0 \ --fastqs=/home/jdoe/mkfastq_pipeline/outs/fastq_path \ --sample=SampleGEX \
This will generate a Loupe Cell Browser .cloupe file, a gene-barcode matrix, and other information.
For more details on running the cellranger count pipeline, consult the
Cell Ranger Gene Expression documentation.
If you generated a Feature Barcoding library alongside the gene expression library, it must be processed at the same time as the gene expression library. Consult the Cell Ranger Feature Barcoding Analysis in the Gene Expression section for more details.
Loupe Analysis
The Loupe Cell Browser and Loupe V(D)J Browser software allow you to tie gene expression and V(D)J
information from these three pipeline runs back together. In Loupe Cell Browser 2.0 and greater, you can import
T Cell and B Cell V(D)J .vloupe files into the Loupe Cell Browser workspace, allowing you to explore
the immune repertoire within indvidual clusters, measure the gene expression of cells within
a clonotype of interest, and create new clusters based on V(D)J data:
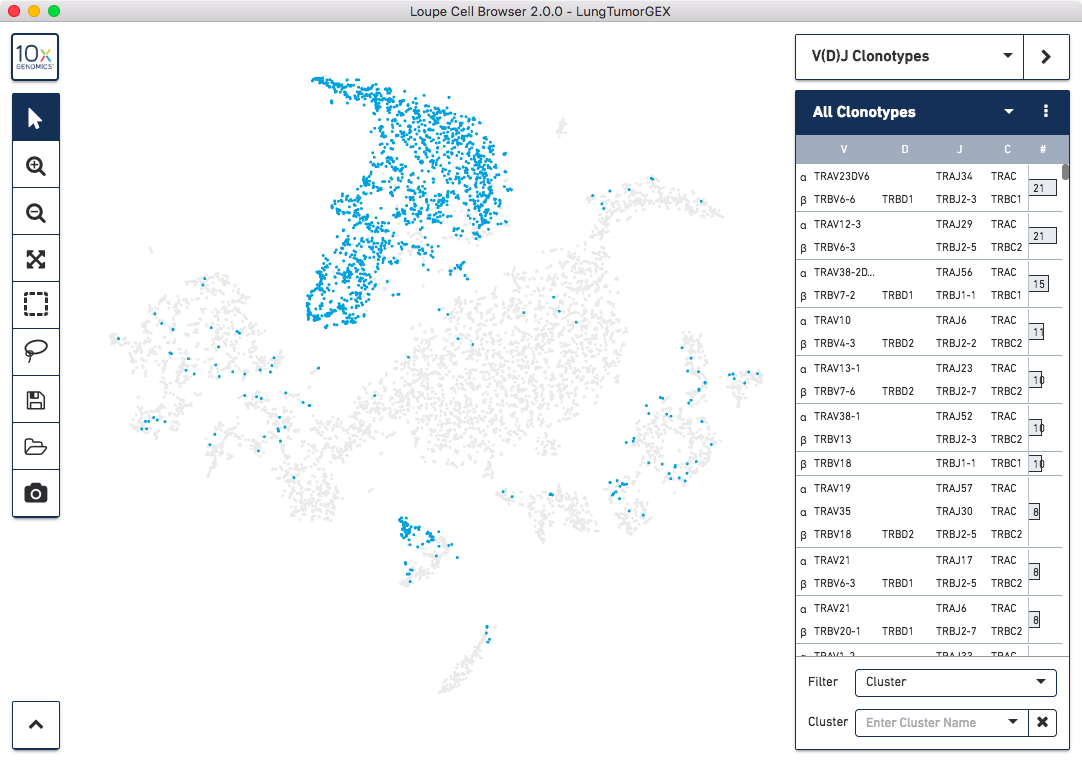
Click here for more information about how to use Loupe Cell Browser for gene expression and V(D)J analysis.
Likewise, you can import gene expression .cloupe files into Loupe V(D)J Browser, in order to
compare clonotypes between gene expression clusters, and explore V(D)J sequences within
clusters in greater detail:
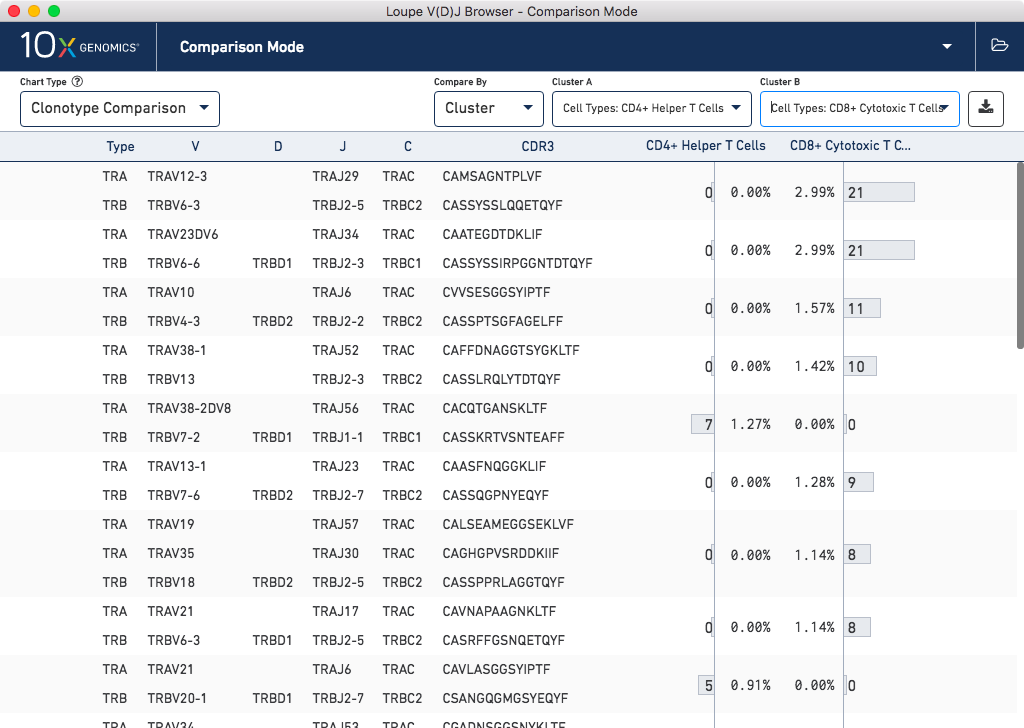
Click here for more information about how to use Loupe V(D)J Browser for more detailed analysis of V(D)J data.
Example Dataset and Tutorial
To learn more about how to conduct multimodal analysis of V(D)J and gene expression data from the same sample, start the Loupe V(D)J + Gene Expression Tutorial. In the tutorial, you will be able to download example 5′ gene expression profiles and T cell clonotypes from a non-small cell lung carcinoma, and use the Loupe browsers to analyze the integrated data.
To learn more about analyzing Feature Barcoding data, start the Immune Profile Analysis with Feature Barcoding and Surface Marker Expression tutorial. In this tutorial, you will download and explore 5′ gene expression and Cell Surface Protein data from a PBMC sample with Loupe Cell Browser.