10x Genomics
Chromium Single Cell Immune Profiling
Cell Ranger3.0, printed on 11/14/2024
Integrated V(D)J and Gene Expression Analysis in Loupe V(D)J Browser
Overview
In this part of the analysis walkthrough, we will use Loupe V(D)J Browser to look at clonotypes within a gene expression cluster, and compare clonotype distributions between clusters. You will need these files to proceed:
- Download Loupe V(D)J Browser 3.0.0
- Download Lung Carcinoma Gene Expression .cloupe file
- Download Lung Carcinoma T Cell .vloupe file
If you're new to Loupe V(D)J Browser, you may want to read the Loupe V(D)J Browser tutorial before continuing.
Loading the Lung Tumor Dataset
To look at clonotypes within gene expression clusters in Loupe V(D)J Browser, you'll need
to first one or more V(D)J .vloupe files, and then import the gene expression .cloupe file.
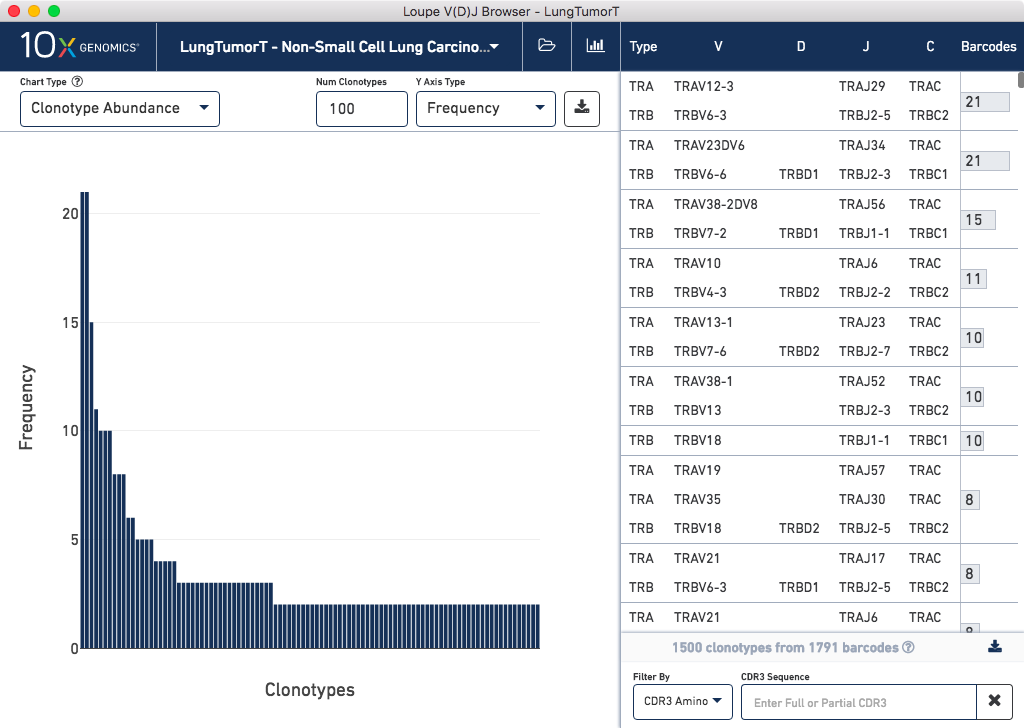
To start, open up Loupe V(D)J Browser, and click on Browse for a Loupe V(D)J Browser File.
Choose the LungTumorT dataset and press Load. You should see the
following after loading:
Next, click on the folder icon on the top toolbar, and select Load clusters from cloupe file.
Choose the LungTumorGEX file that you either downloaded from the previous
exercise, or from the link above.
After importing gene expression data, you will see an icon next to any V(D)J samples for which
corresponding gene expression data has been loaded. You should see the icon next to the
LungTumorT sample:
The icon signals that you can filter clonotypes in that file by cluster, and use Comparison Mode to compare clonotypes between gene expression clusters.
Cluster Clonotypes in Depth
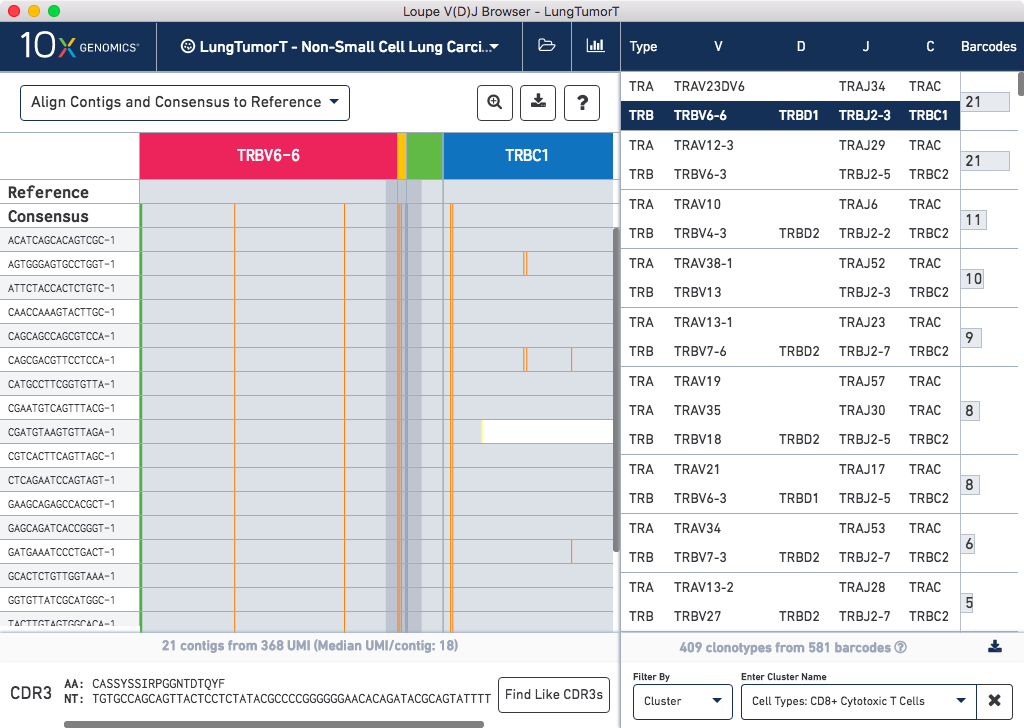
Select the Cluster option from the Filter By field below the clonotype list. Type 'CD8' in the Enter Cluster Name box. The CD8+ Cytotoxic T Cells cluster will appear from the autocomplete list. Select it to limit the clonotype list to cells in that cluster. Clicking on each chain in the list will show you the mutations (vertical lines) against the combined V(D)J reference for the chain.
Next, let's compare clonotype distributions between gene expression clusters.
Clonotype Comparison Between Gene Expression Clusters
Next, let's use Loupe V(D)J Browser to identify any clonotypes that are unique to the CD4+ Helper T Cell cluster and the CD8+ Cytotoxic T Cell cluster.
Select Comparison Mode from the file selector, and then choose Clonotype Comparison from the Chart Type chart selector above the sample table. Since you have already loaded gene expression data, you may choose Cluster from the Compare By dropdown. Under Cluster A, enter "CD4+ Helper T Cells", and under Cluster B, enter "CD8+ Cytotoxic T Cells":
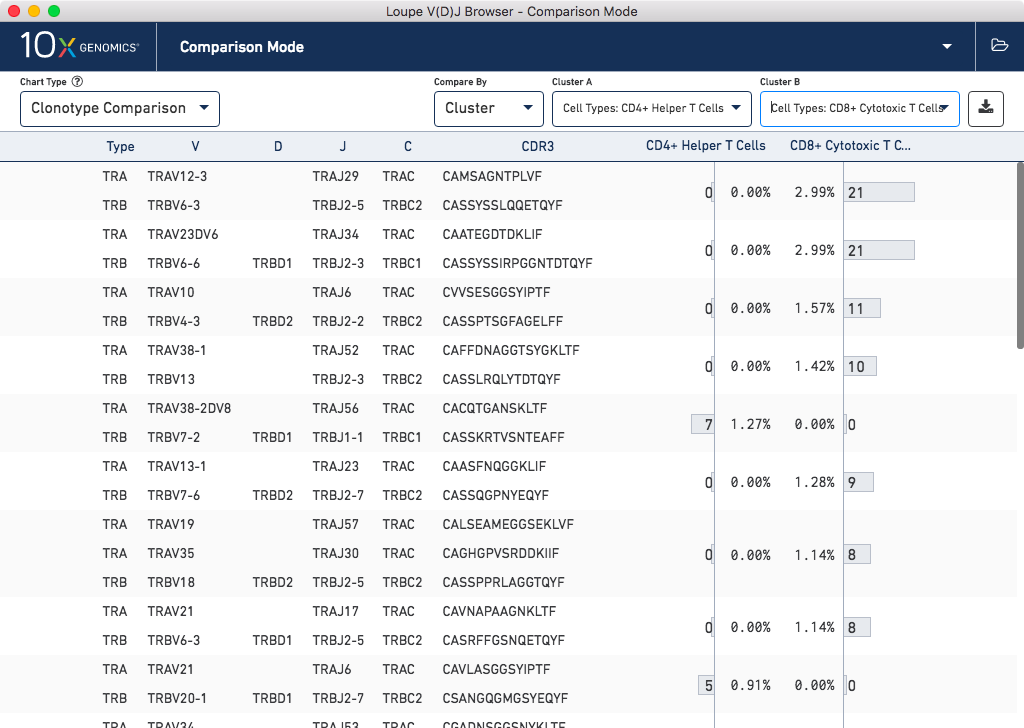
You can see that there are a set of clonotypes which are unique to the CD8+ Cytotoxic T Cell cluster, and another set that are unique to the CD4+ Helper T Cell cluster. Clonotype differences are ordered by p-value, as computed by applying Fisher's exact test. To explore a clonotype in more detail, you can copy a CDR3 sequence by clicking on it, and using the CDR3 Amino filter in the single-sample clonotype/chain view. Finally, you can export the list of differing clonotypes to a CSV file, for further downstream processing.
Next Steps
Now that you've seen how to combine V(D)J and gene expression data with Loupe Cell Browser and Loupe V(D)J Browser, you'll have ideas about how to use the Chromium™ Single Cell V(D)J Solution for your own experiments. If you have any questions or feedback about software or other parts of the integrated V(D)J and gene expression workflow, please contact support@10xgenomics.com.