10x Genomics
Chromium Single Cell Gene Expression
Cell Ranger6.1, printed on 07/11/2025
Web Summary
The cellranger count pipeline outputs an interactive summary HTML file named web_summary.html that contains summary metrics and automated secondary analysis results. If an issue was detected during the pipeline run, an alert appears on this page. More information about alerts is provided in the Cell Ranger troubleshooting documentation.
Table of Contents
Summary view
The run summary from cellranger count can be viewed by clicking Summary in the top left tab of the HTML file. The summary metrics describe sequencing quality and various characteristics of the detected cells. Similar web summaries are also output from the cellranger reanalyze and cellranger aggr pipelines.
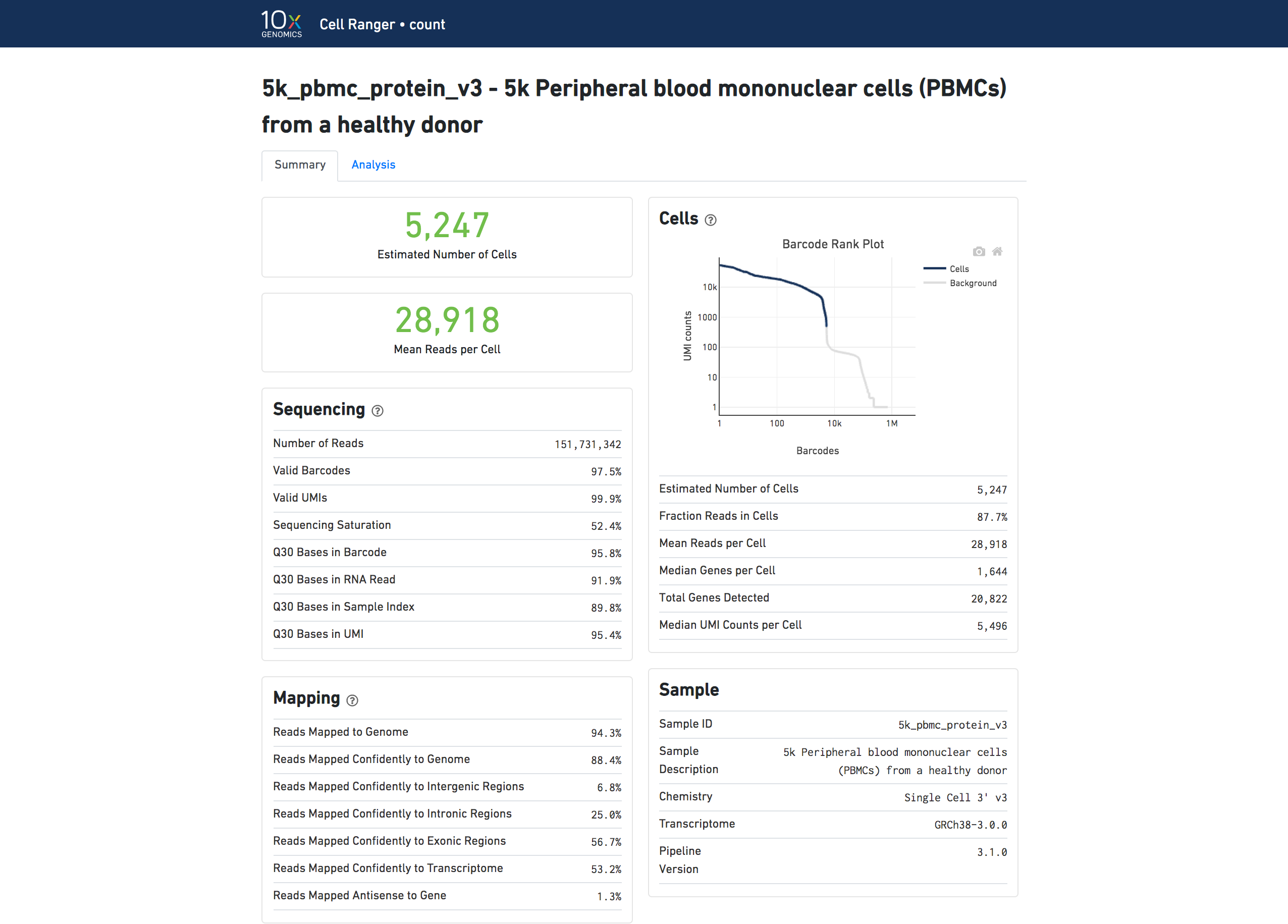
The number of cells detected, the mean reads per cell, and the median genes detected per cell are prominently displayed near the top of the page.
Click the ? icons next to Sequencing, Mapping, and Cells sections to display information about each metric in the dashboard.
The Barcode Rank Plot under the Cells dashboard shows the distribution of barcode counts and which barcodes were inferred to be associated with cells. The y-axis is the number of UMI counts mapped to each barcode and the x-axis is the number of barcodes below that value. A steep drop-off is indicative of good separation between the cell-associated barcodes and the barcodes associated with empty partitions. Since barcodes can be associated with cells based on their UMI count or by their RNA profiles, some regions of the graph can contain both cell-associated and background-associated barcodes. The color of the graph represents the local density of barcodes that are cell-associated. See Cell Calling Algorithms for more details.
Analysis view
The automated secondary analysis results can be viewed by clicking the Analysis tab in the top left corner. Click the ? icons next to each section title to display information about the secondary analyses displayed in the dashboard.
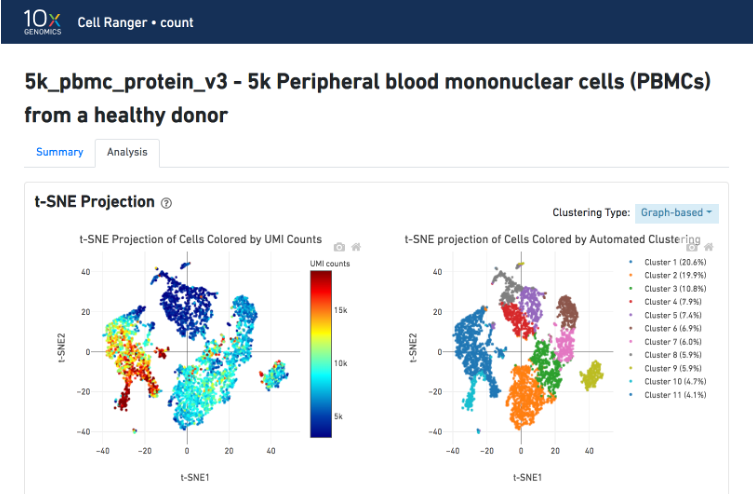
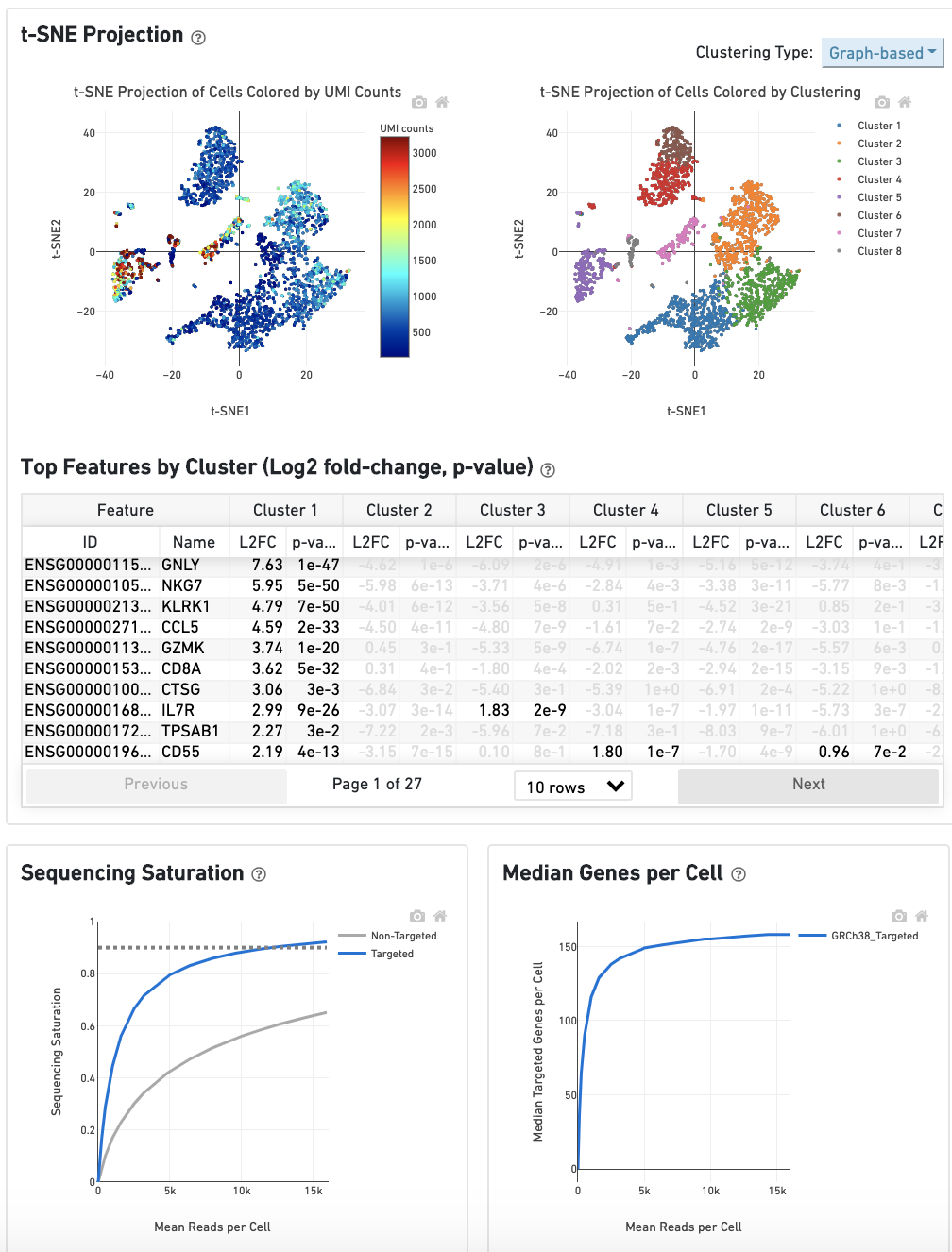
The t-SNE Projection section shows the data reduced to two dimensions, colored by UMI count (left) or clustering (right). It is a good starting point to explore structure in the data. The projection colored by UMI counts is indicative of the RNA content of the cells and often correlates with cell size - redder points are cells with more RNA in them. For the projection colored by clustering results, select the type of clustering analysis to display from the drop-down button on the upper right (Graph-based by default) - change this to vary the type of clustering and/or number of clusters (K=2-10) that are assigned to the data.
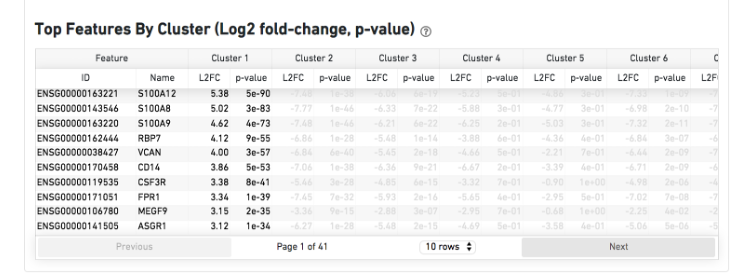
The Top Features By Cluster table shows which genes are differentially expressed in each cluster relative to all other clusters (Graph-based by default). To find the genes associated with a particular cluster, click the L2FC or p-value column headers associated with a given cluster number to sort the table by specificity for that cluster.
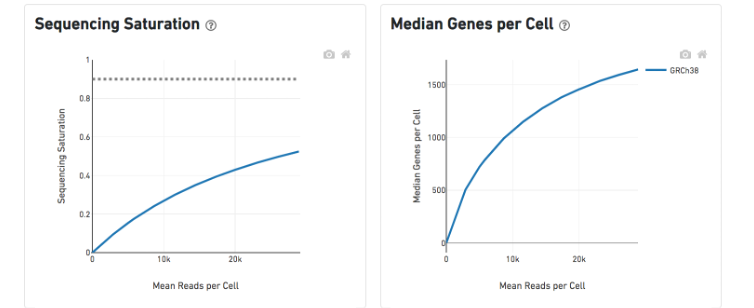
The Sequencing Saturation plot shows the effect of decreased sequencing depth on sequencing saturation, which is a measure of the fraction of library complexity that was observed. The right-most point on the line is the full sequencing depth obtained in this run.
Similarly, the Median Genes per Cell plot shows the effect of decreased sequencing depth on median genes per cell, which is a way of measuring data yield as a function of depth. The right-most point on the line is the full sequencing depth obtained in this run.
Feature Barcode view
If you use Cell Ranger to process Feature Barcode data, new metrics specific to Feature Barcode libraries will appear in the Summary tab in a new dashboard (Antibody Application, CRISPR Application).
Below is an example web summary for Gene Expression and Antibody Capture libraries:
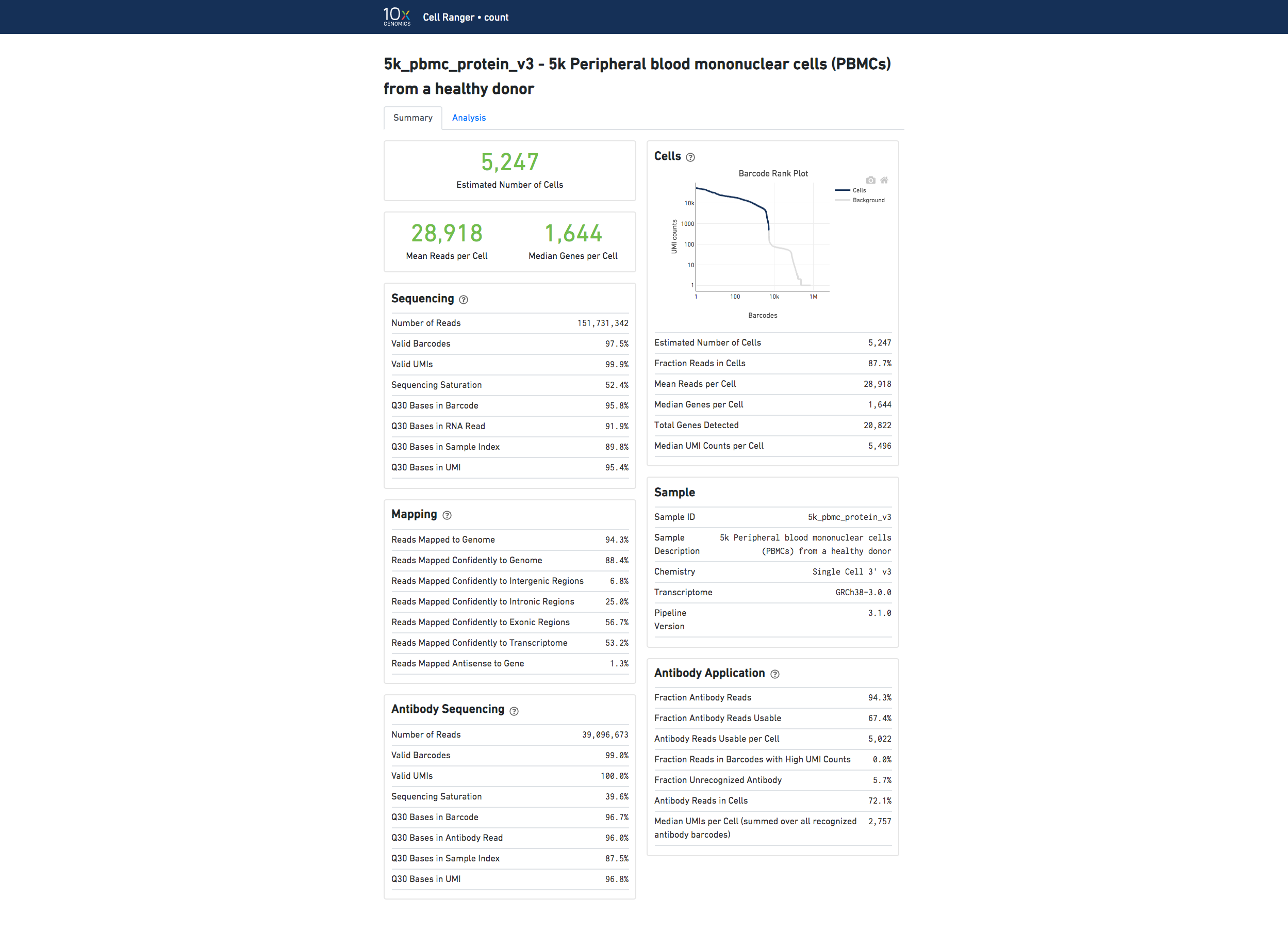
For information about the web summary for an Antibody Capture only analysis, go here.
Here is an example web summary for Gene Expression and CRISPR Guide Capture libraries:
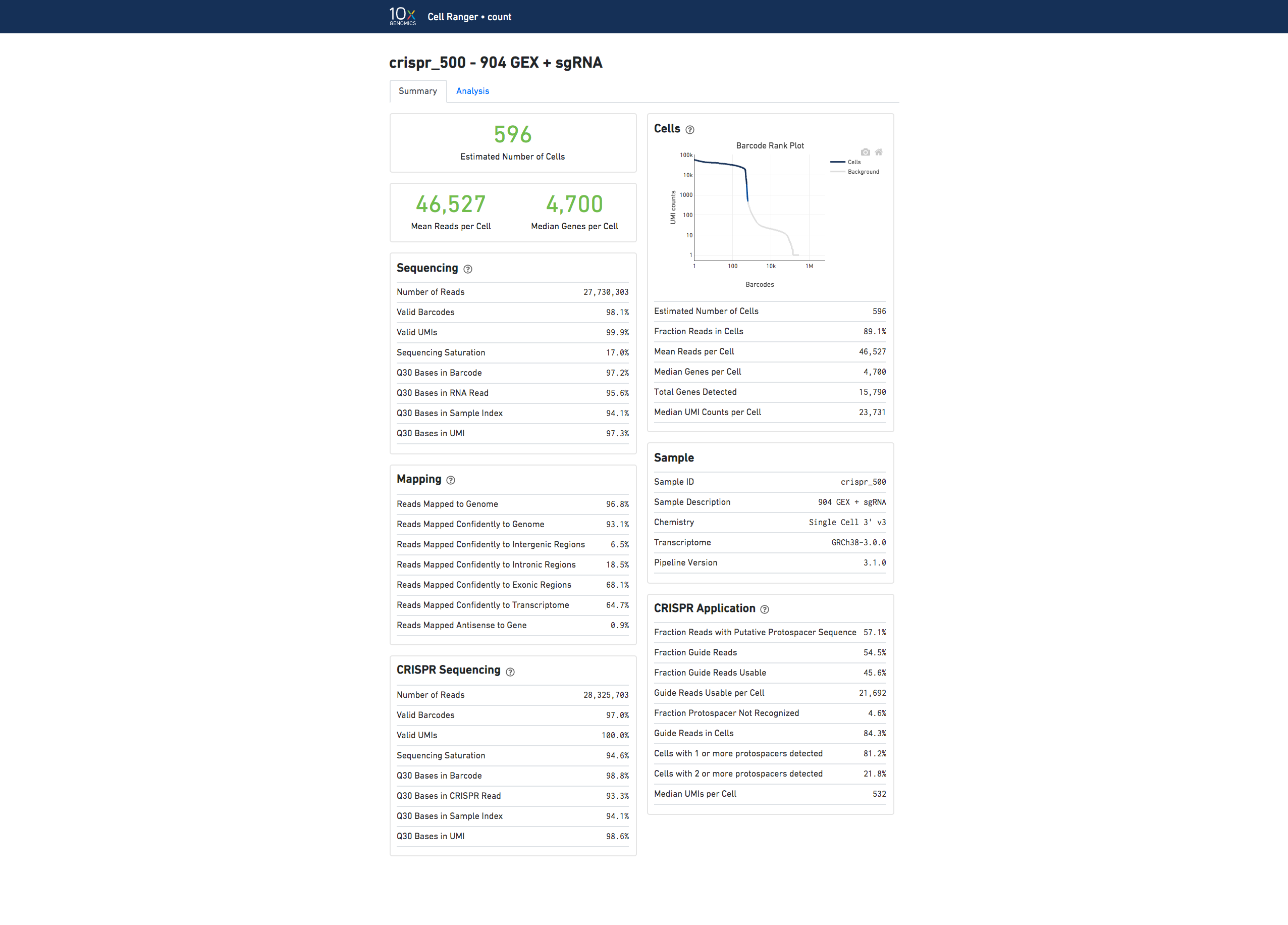
The Analysis tab view for Feature Barcode experiments contains the same secondary analysis as gene expression only experiments, and uses only the gene expression part of the feature-barcode matrix for dimensionality reduction, clustering, and differential gene expression analysis.
Multiple species
If you analyzed an experiment with data from multiple species, the output will look different. For example, this human-mouse mixing experiment consists of a mix of approximately 500 human (HEK293T) cells and 500 mouse (3T3) cells in a 1:1 ratio.
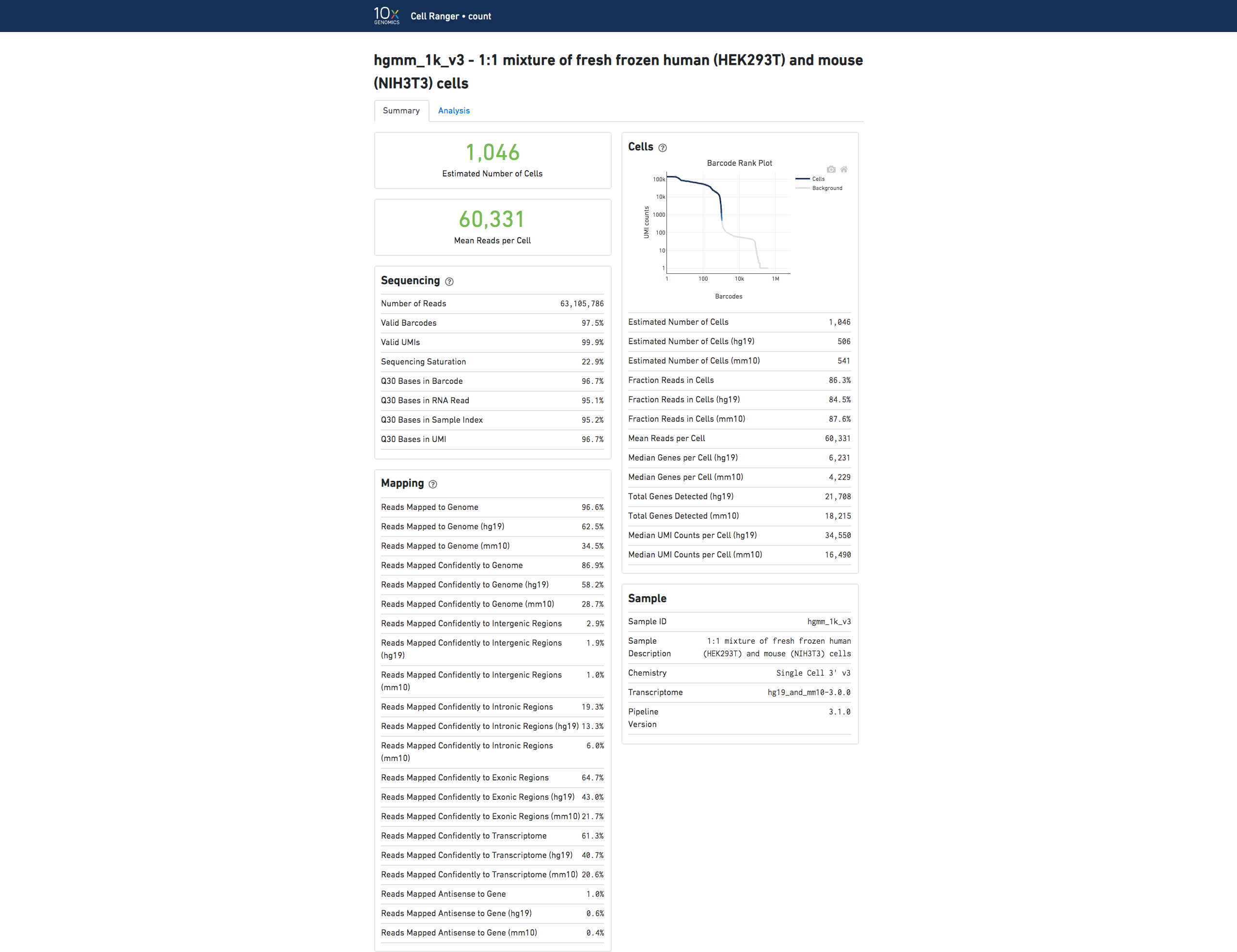
Click the ? at the top of each dashboard to display information about each metric.
In the Analysis view, running the human-mouse mixing experiment results in the following GEM Partitions plot:
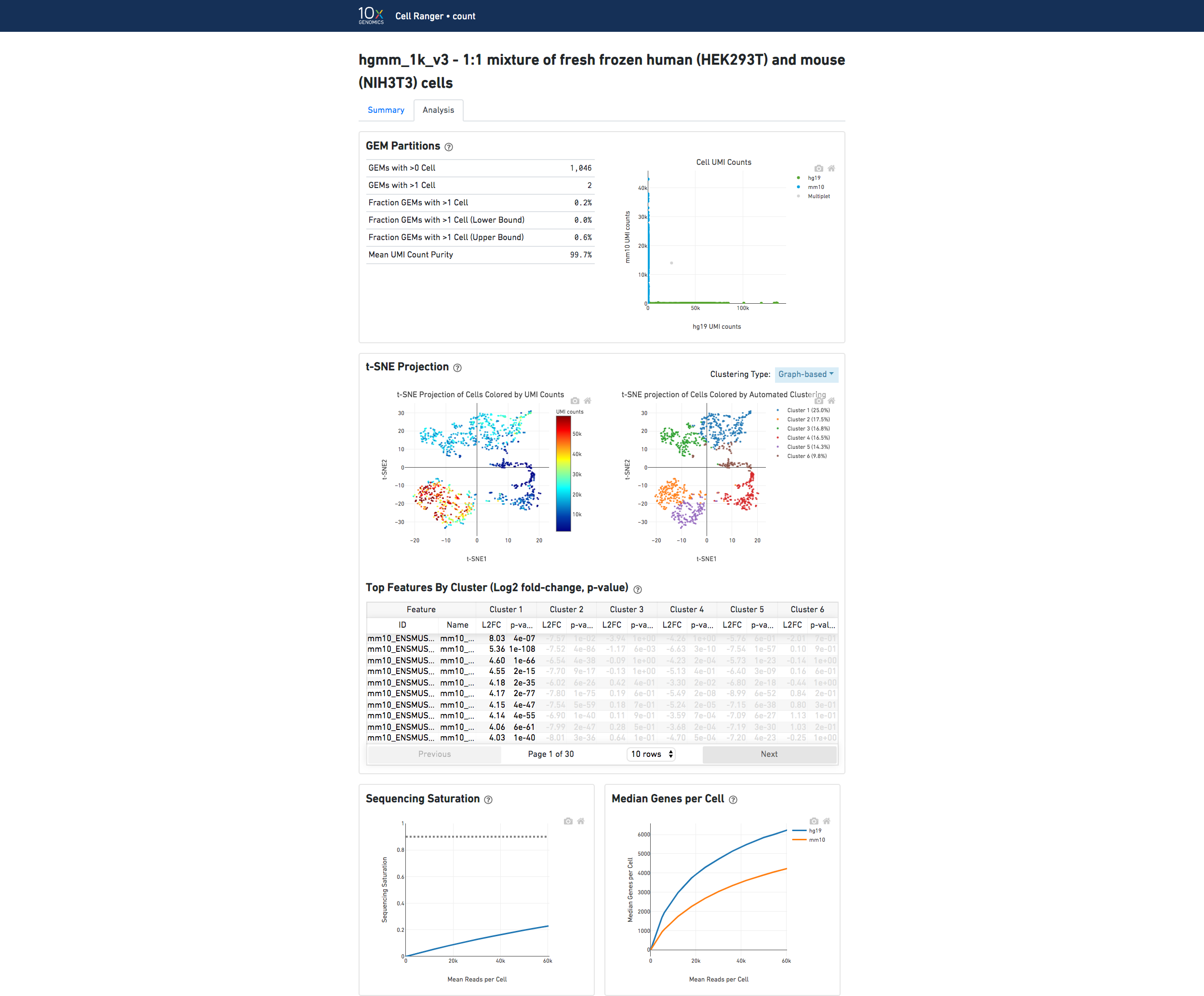
Each point represents a barcode. The gray points are inferred to be associated with GEM partitions that contained more than one cell because they contained a large number of molecules that came from human cells and mouse cells. The GEMs containing human-human and mouse-mouse cell combinations are not visible here, but their presence is inferred in the Fraction GEMs with >1 Cell metric shown next to the plot.
Targeted Gene Expression view
If you ran a Targeted Gene Expression experiment, the output includes modifications that highlight targeting performance. The following is an example of the Summary tab from a cellranger count targeted run.

The Analysis tab includes a Targeted Enrichment dashboard with metrics that assess targeting performance. On the right is a scatterplot of read counts per gene versus UMI counts per gene. A successful targeted experiment where targeted genes are well-enriched looks like the one below, with targeted genes (blue) cleanly separated from non-targeted genes (gray). See Targeted Gene Expression Algorithms for more information on how gene enrichments are computed.
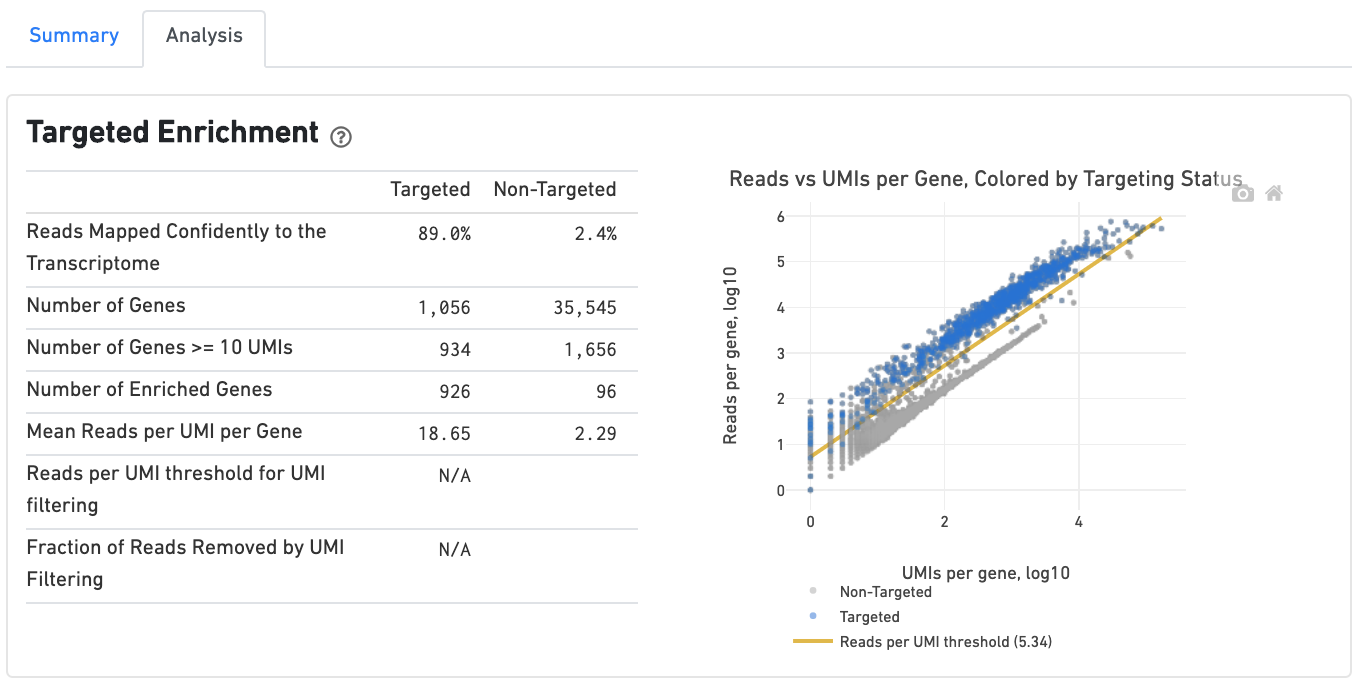
The remainder of the Analysis tab is very similar to that of a regular gene expression analysis, but focuses on targeted genes (Sequencing Saturation, Median Genes per Cell); all secondary analysis (t-SNE, differential expression) is done using only targeted genes.
Do you have questions or feedback about this documentation? Please contact support@10xgenomics.com.