10x Genomics
Chromium Single Cell Gene Expression
Cell Ranger2.1, printed on 08/12/2025
What is Cell Ranger?
Cell Ranger is a set of analysis pipelines that process Chromium single-cell RNA-seq output to align reads, generate gene-cell matrices and perform clustering and gene expression analysis. Cell Ranger includes four pipelines relevant to single-cell gene expression experiments:
-
cellranger mkfastq demultiplexes raw base call (BCL) files generated by Illumina sequencers into FASTQ files. It is a wrapper around Illumina's bcl2fastq, with additional useful features that are specific to 10x libraries and a simplified sample sheet format.
-
cellranger count takes FASTQ files from cellranger mkfastq and performs alignment, filtering, barcode counting, and UMI counting. It uses the Chromium cellular barcodes to generate gene-barcode matrices, determine clusters, and perform gene expression analysis. The count pipeline can take input from multiple sequencing runs on the same library.
-
cellranger aggr aggregates outputs from multiple runs of cellranger count, normalizing those runs to the same sequencing depth and then recomputing the gene-barcode matrices and analysis on the combined data. The aggr pipeline can be used to combine data from multiple samples into an experiment-wide gene-barcode matrix and analysis.
-
cellranger reanalyze takes gene-barcode matrices produced by cellranger count or cellranger aggr and reruns the dimensionality reduction, clustering, and gene expression algorithms using tunable parameter settings.
These pipelines combine Chromium-specific algorithms with the widely used RNA-seq aligner STAR. Output is delivered in standard BAM, MEX, CSV, HDF5 and HTML formats that are augmented with cellular information.
Terminology
Throughout the documentation, you will see references to samples, libraries and sequencing runs. We define these as follows:
- Sample: A cell suspension extracted from a single biological source (blood, tissue, etc).
- Library: A 10x-barcoded sequencing library prepared from a single sample, corresponding to a single chip channel of a 10x Chromium Controller run.
- Sequencing Run (or Flowcell): A flowcell containing data from one sequencing instrument run. The sequencing data can be further addressed by lane and by one or more sample indices.
The relationship between these terms can be complex:
- A single sample may be prepared into multiple libraries; this is commonly done to achieve higher cell counts without overloading a single 10x Chromium Controller run.
- A single library may be sequenced across multiple flowcells, and then combined as if all the reads came from one sequencing run.
- A single flowcell may contain multiple libraries, separated using different lanes, sample indices, or both.
- The definition of "library" makes no distinction between two channels of the same physical 10x chip, versus the same channel of two different chips.
Chemistry
Cell Ranger 1.2 and later support libraries generated by the Chromium Single Cell 3′ v1 and v2 reagent kits, whereas Cell Ranger 1.1 and earlier do not support v2 libraries. Support for 5′ reagents is new in Cell Ranger 2.1.
Workflows
The Cell Ranger workflow starts with demultiplexing the raw base call (BCL) files for each flowcell directory. 10x recommends using cellranger mkfastq as described in Generating FASTQs. If you are starting with FASTQ files demultiplexed with bcl2fastq directly, or from a public source such as SRA, skip mkfastq and begin with cellranger count. Please see the Specifying Input FASTQs page for specific guidelines on which arguments to use for your scenario.
The subsequent steps vary depending on how many samples, libraries, and flowcells you have. We will describe them in order of increasing complexity:
Single Sample, Library, and Flowcell
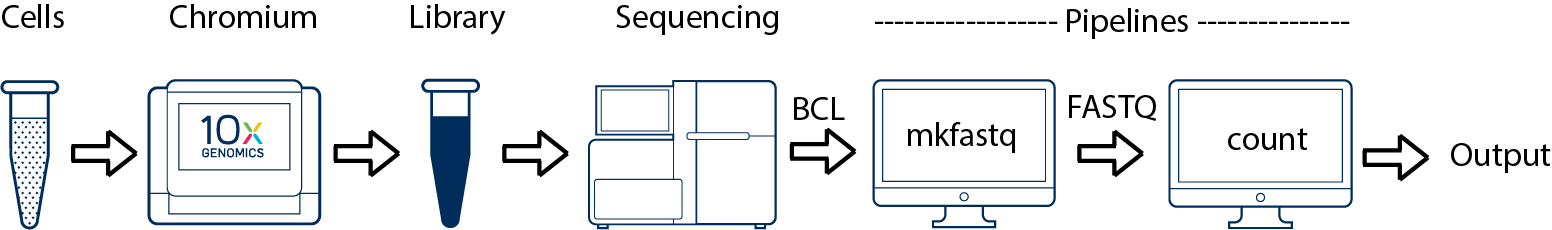
This is the simplest scenario. If you have a single biological sample corresponding to a single library and sequenced on a single flowcell, you would generate FASTQs using cellranger mkfastq, and run cellranger count as described in Single-Library Analysis.
One Library, Multiple Flowcells
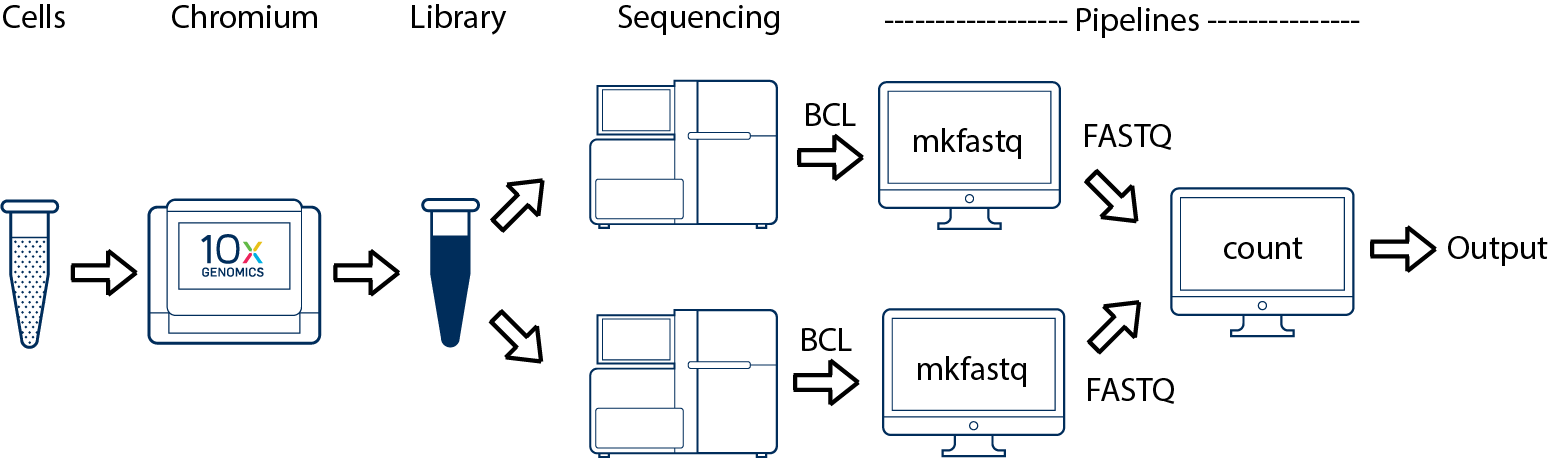
If you have a library which was sequenced across multiple flowcells (for example to increase sequencing saturation), all of the reads can be combined in a single instance of the cellranger count pipeline. This process is described in Specifying Input Fastqs.
One Sample, Multiple Libraries
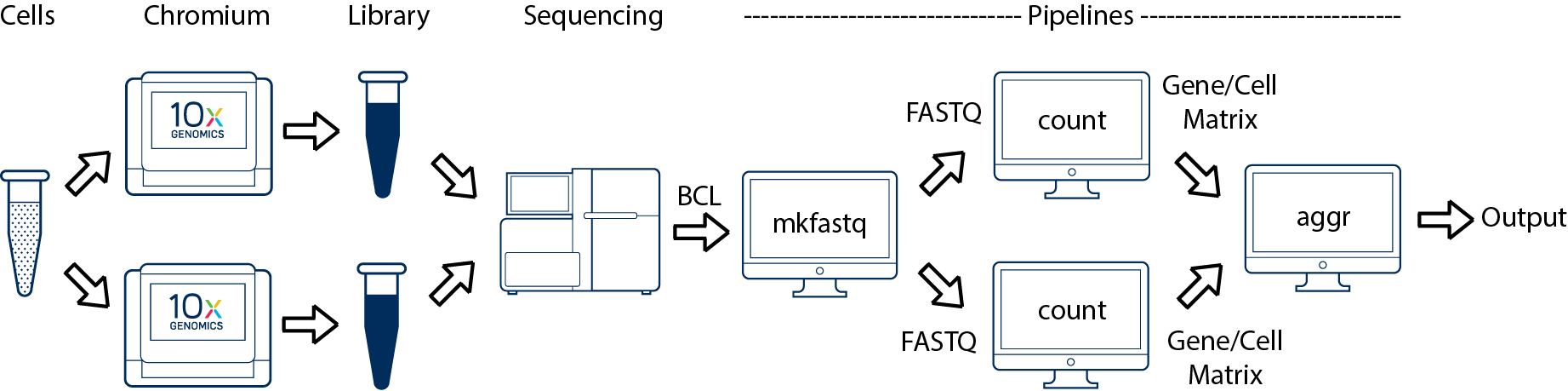
This comprises two scenarios.
Technical Replicates: If you prepared multiple libraries (technical replicates) from the same sample and want to treat them as distinct samples, then each one should be run through a separate instance of cellranger count. Once those are completed, you can perform a combined analysis using cellranger aggr, as described in Multi-Library Aggregation. (This is illustrated in the figure).
Pooled Libraries: If you prepared multiple libraries from the same sample and want to pool them and analyze the combined data as a single sample, then you will need to use the MRO syntax to treat the multiple libraries as coming from a single sample. (This is not illustrated in the figure).
Multiple Biological Samples
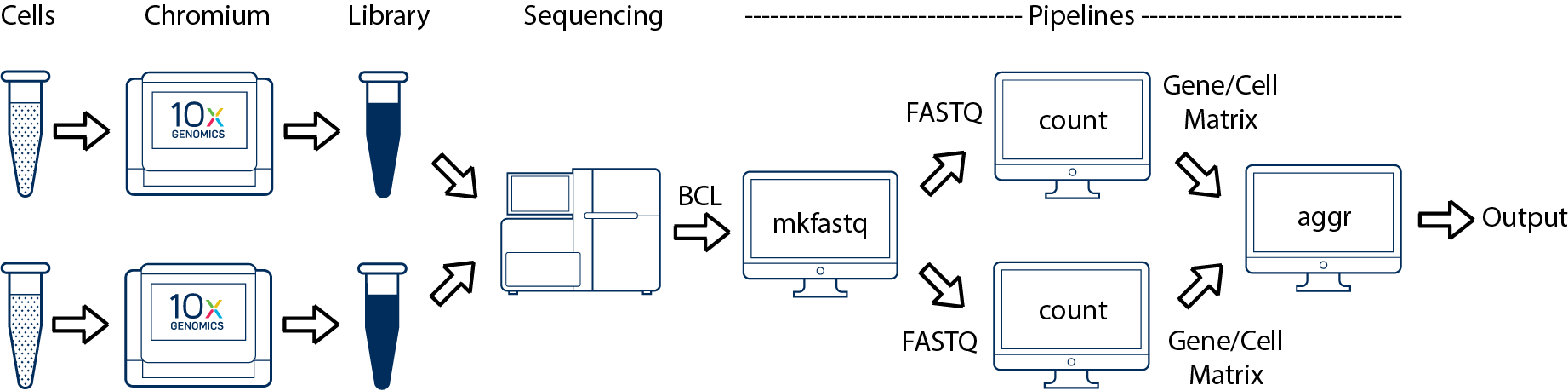
For an experiment involving multiple biological samples, you must run cellranger count separately for each individual library deriving from each of those samples. For instance, if your experiment involves two samples then you will have to run cellranger count two times. Then you can aggregate them with a single instance of cellranger aggr.
One or more 5′ Gene Expression and V(D)J Enriched Libraries from the Same Biological Sample
5′ gene expression libraries and V(D)J enriched libraries generated from the same cDNA product must be processed by cellranger count and cellranger vdj respectively. Refer to Single Cell V(D)J + 5′ Gene Expression for more information.