10x Genomics
Chromium Single Cell CNV
Cell Ranger DNA1.1 (latest), printed on 07/29/2025
Loupe scDNA Browser: Trace
| Analysis software for the 10x Genomics single cell DNA product is no longer supported. Raw data processing pipelines and visualization tools are available for download and can be used for analyzing legacy data from 10x Genomics kits in accordance with our end user licensing agreement without support. |
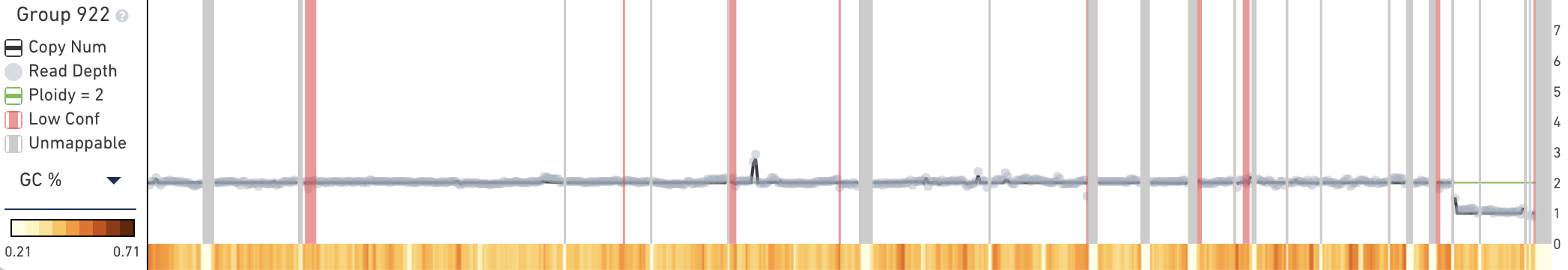
The trace displays copy number, read depth, and other data about the current subtree root. When you first open the MKN-45 tutorial dataset, the trace will contain information about the root node of the entire cell hierarchy, labeled Group 922. The trace, in contrast to the heatmap, shows data about the currently selected group or cell, not data about the descendants of the selected node.
The trace displays the following data for the current root group or cell:
- Copy Number as a line; the scale is shown to the right of the plot
- Read Depth as gray dots, scaled to approximate reads per megabase
- Low Confidence Regions shown as red overlays
- Unmappable Regions shown as gray rectangles
- A green line representing the current white point in the heatmap
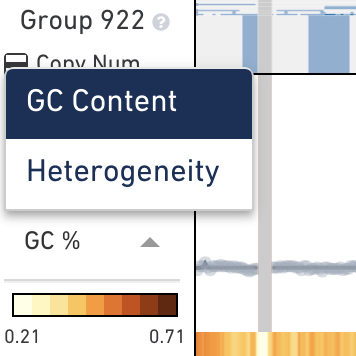
Below the main plot is a colored strip which, by default, displays the GC percentage of copy number calls at this node. To switch this colored strip to display heterogeneity, click on the trace colorbar selector and select Heterogeneity. Remember that heterogeneity is only defined for groups of cells and is undefined (gray) for single cells.
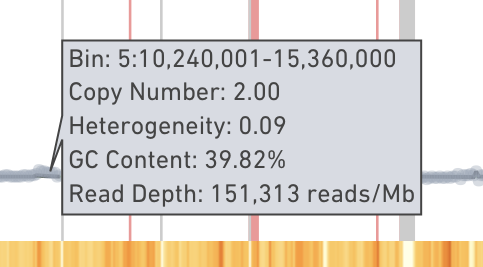
Hovering on the plot displays the exact numbers of aforementioned data: genomic range of the bin, ploidy, heterogeneity, GC content, and read depth.
- 1.0
- Loupe scDNA Browser v1.1 (latest)