10x Genomics
Chromium Single Cell Multiome ATAC + Gene Exp.
Cell Ranger ARC1.0, printed on 07/09/2025
Generating FASTQs with cellranger-arc mkfastq
Table of Contents
- Overview
- Example Workflows
- Arguments and Options
- Example Data
- Running mkfastq with a simple CSV samplesheet
- Running mkfastq with an Illumina Experiment Manager sample sheet
- Checking FASTQ output
- Reading Quality Control Metrics
- Troubleshooting
Overview
The cellranger-arc workflow starts by demultiplexing the
Illumina sequencer's base call files (BCLs) for each flow cell directory (ATAC
or GEX) into FASTQ files. 10x has developed cellranger-arc
mkfastq, a pipeline that wraps Illumina's bcl2fastq and provides a
number of convenient features in addition to the features of bcl2fastq:
- Supports demultiplexing of ATAC or GEX flow cells
- Translates 10x sample index names into the corresponding oligonucleotides in the sample index.
- Supports a simplified CSV samplesheet format to handle 10x use cases.
- Generates sequencing and 10x-specific quality control metrics, including barcode quality, accuracy, and diversity.
- Supports most
bcl2fastqarguments, such as--use-bases-mask.
The Multiome ATAC library is single-indexed while the Multiome GEX library is
dual-indexed. cellranger-arc mkfastq can auto-detect the
type of flow cell based on the length of the I2 read and selects the appropriate
mode depending on the sample indexes used, and enables index-hopping filtering
automatically for dual-indexed flow cells. For example, a Multiome GEX library
prepared with the Dual Index Kit TT Set A, well A1 can be specified in the
samplesheet as SI-TT-A1, and cellranger-arc mkfastq will
recognize the i7 and i5 indices as GTAACATGCG and AGTGTTACCT, respectively.
Similarly for a Multiome ATAC library prepared with Single Index Kit N Set A,
well A1 can be specified in the samplesheet as SI-NA-A1, and
cellranger-arc mkfastq will recognize the four i7 indexes
AAACGGCG, CCTACCAT, GGCGTTTC, and TTGTAAGA and merge the resulting FASTQ
files.
Example Workflows
The compute workflow begins with running one instance of cellranger-arc mkfastq for each flow cell of data being analyzed. The same command cellranger-arc mkfastq can be used to demultiplex ATAC and GEX flow cells. Once the ATAC flow cell(s) and GEX flow cell(s) are successfully demultiplexed, we run one instance of cellranger-arc count for each paired Multiome ATAC and GEX library; independent of the number of sequencing runs of each library. We illustrate the above using specific examples below.
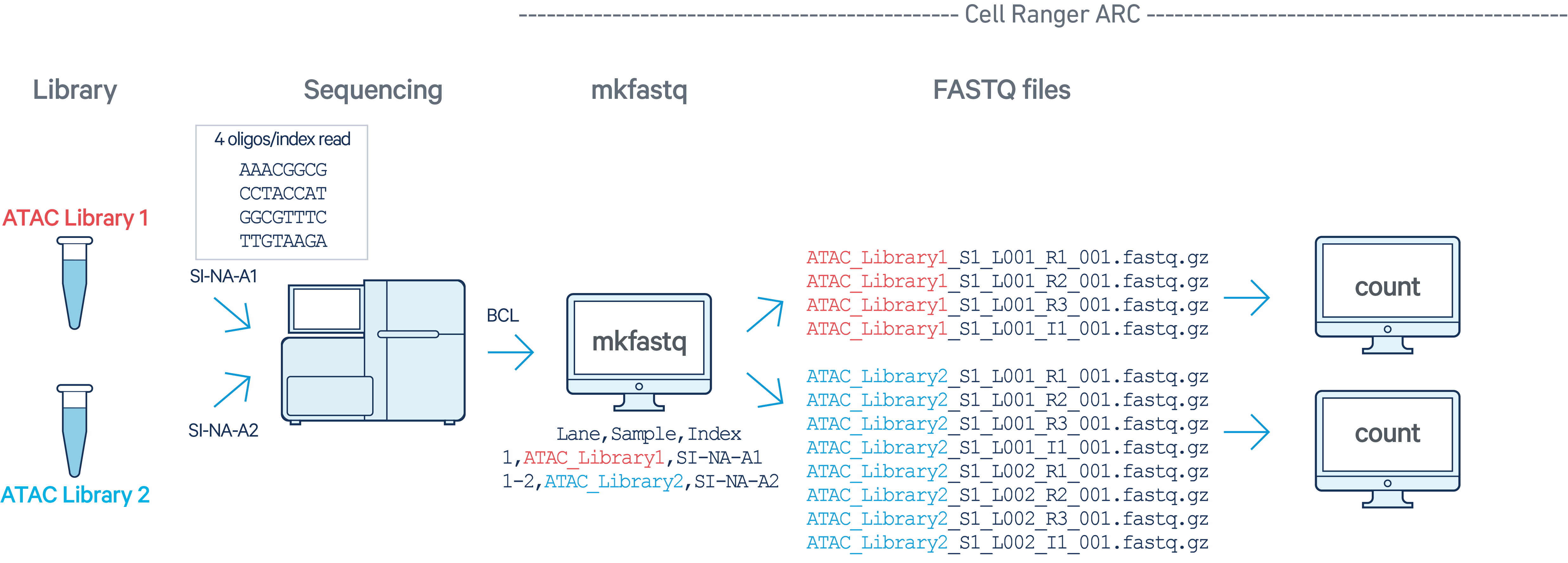
ATAC
In this example, we have two Multiome ATAC libraries (each processed through a
separate Chromium chip channel with sample indices SI-NA-A1 and SI-NA-A2)
that are multiplexed on a single flow cell. Note that after running
cellranger-arc mkfastq, we run a separate instance of
cellranger-arc count on each library:
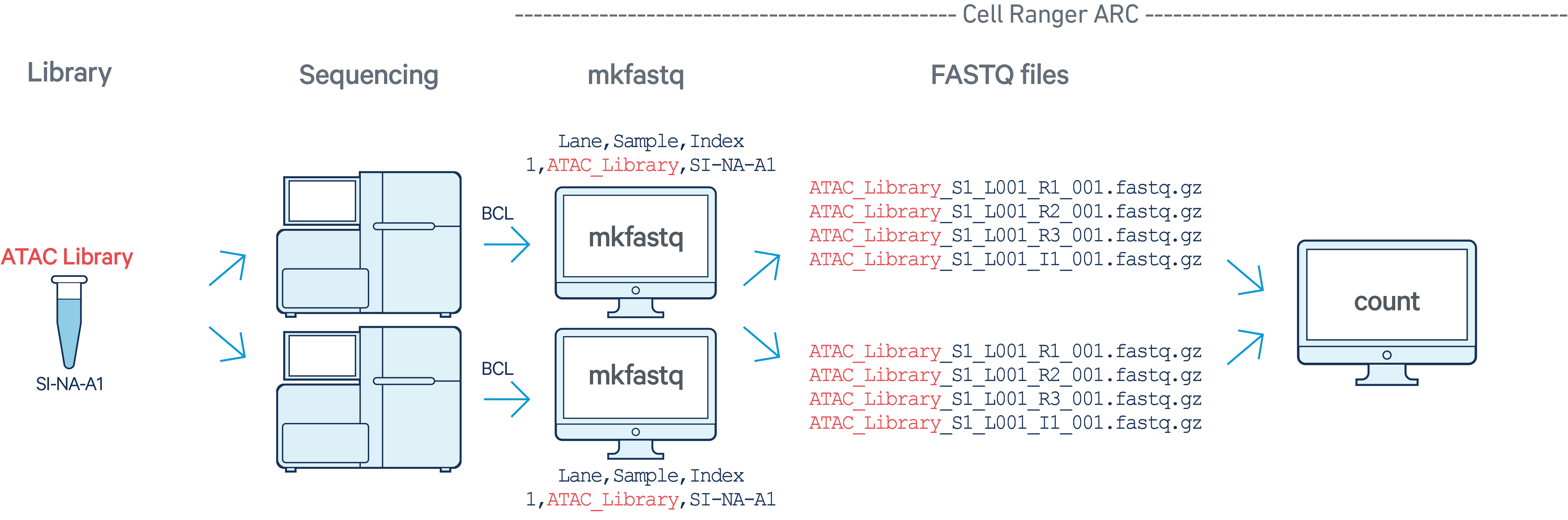
In this example, we have one Multiome ATAC library with sample index SI-NA-A1
sequenced on two flow cells. Note that after running
cellranger-arc mkfastq, we run a single instance of the
pipeline on all the FASTQ files generated:
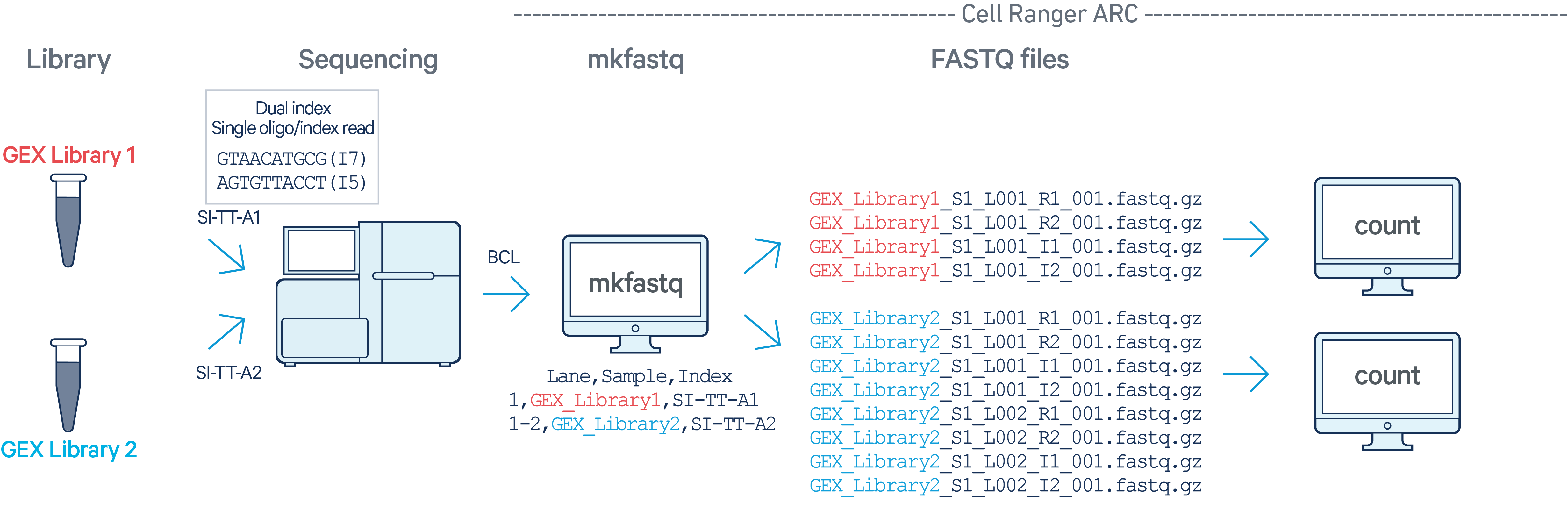
GEX
In this example, we have two Multiome GEX libraries (each processed through a
separate Chromium chip channel with sample indices SI-TT-A1 and SI-TT-A2)
that are multiplexed on a single flow cell. Note that after running
cellranger-arc mkfastq, we run a separate instance of
cellranger-arc count on each library:
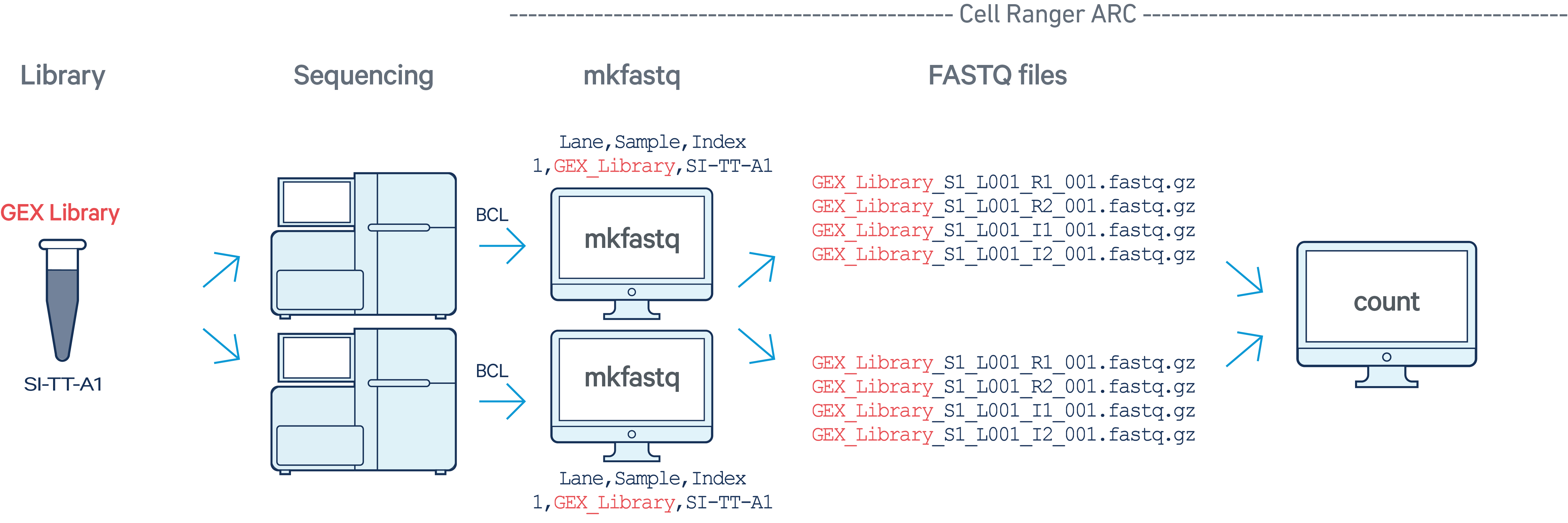
In this example, we have one Multiome GEX library with sample index SI-TT-A1
sequenced on two flow cells. Note that after running
cellranger-arc mkfastq, we run a single instance of the
pipeline on all the FASTQ files generated:
Arguments and Options
cellranger-arc mkfastq accepts additional options beyond
those shown in the table below because it is a wrapper around
bcl2fastq. Consult the User Guide for Illumina's
bcl2fastq
for more information.
| Parameter | Function |
|---|---|
| --run | (Required) The path of Illumina BCL run folder. |
| --id | (Optional; defaults to the name of the flow cell referred to by --run) Name of the folder created by mkfastq. |
| --samplesheet | (Optional) Path to an Illumina Experiment Manager-compatible sample sheet which contains 10x sample index names (e.g., SI-NA-A1 or SI-TT-A12) in the sample index column. All other information, such as sample names and lanes, should be in the sample sheet. |
| --sample-sheet | (Optional) Equivalent to --samplesheet above. |
| --csv | (Optional) Path to a simple CSV with lane, sample, and index columns, which describe the way to demultiplex the flow cell. The index column should contain a 10x sample dual-index name (e.g., SI-TT-A12). This is an alternative to the Illumina IEM sample sheet, and will be ignored if --samplesheet is specified. |
| --simple-csv | (Optional) Equivalent to --csv above. |
| --filter-dual-index | (Optional) Only demultiplex samples identified by i7/i5 dual-indices (e.g., SI-TT-A6), ignoring single-index samples. Single-index samples will not be demultiplexed. Also notice that cellranger-arc will run single-index data, but it is not supported. |
| --qc | (Optional) Calculate both sequencing and 10x-specific metrics, including per-sample barcode matching rate. Will not be performed unless this flag is specified. Not supported for NovaSeq flow cells. |
| --lanes | (bcl2fastq option) Comma-delimited series of lanes to demultiplex (e.g. 1,3). Use this if you have a sample sheet for an entire flow cell but only want to generate a few lanes for further 10x analysis. |
| --use-bases-mask | (bcl2fastq option) Same meaning as for bcl2fastq. Use to clip extra bases off a read if you ran extra cycles for QC. |
| --delete-undetermined | (bcl2fastq option) Delete the Undetermined FASTQs generated by bcl2fastq. Useful if you are demultiplexing a small number of samples from a large flow cell. |
| --output-dir | (bcl2fastq option) Generate FASTQ output in a path of your own choosing, instead of flow_cell_id/outs/fastq_path. |
| --project | (bcl2fastq option) Custom project name, to override the samplesheet or to use in conjunction with the --csv argument. |
| --jobmode | (Martian option) Job manager to use. Valid options: local (default), sge, lsf, slurm or a .template file. |
| --localcores | (Martian option) Set max cores the pipeline may request at one time. Only applies when --jobmode=local. |
| --localmem | (Martian option) Set max GB the pipeline may request at one time. Only applies when --jobmode=local. |
Example Data
cellranger-arc mkfastq recognizes two file formats for
describing samples: a simple, three-column CSV format, and the Illumina
Experiment Manager (IEM) sample sheet format used by bcl2fastq. We
illustrate these formats with a Multiome ATAC flow cell and Multiome GEX flow
cell example.
To follow along, do the following:
- Download the tiny-bcl-atac tar file and tiny-bcl-gex tar file.
- Untar both the cellranger-arc-tiny-bcl-atac-1.0.0.tar.gz and cellranger-arc-tiny-bcl-gex-1.0.0.tar.gz tar files in a convenient location.
- Download the simple CSV layout files: cellranger-arc-tiny-bcl-atac-simple-1.0.0.csv and cellranger-arc-tiny-bcl-gex-simple-1.0.0.csv.
- Download the Illumina Experiment Manager sample sheets: cellranger-arc-tiny-bcl-atac-samplesheet-1.0.0.csv and cellranger-arc-tiny-bcl-gex-samplesheet-1.0.0.csv.
Running mkfastq with a Simple CSV Samplesheet
A simple csv samplesheet is recommended for most sequencing experiments.
The simple csv format has only three columns (Lane, Sample, Index), and is thus
less prone to formatting errors. You can see an example of this in
cellranger-arc-tiny-bcl-atac-simple-1.0.0.csv:
Lane,Sample,Index 1,test_sample_atac,SI-NA-A1
and in cellranger-arc-tiny-bcl-gex-simple-1.0.0.csv:
Lane,Sample,Index 1,test_sample_gex,SI-TT-A1
Here are the options for each column:
| Lane | Which lane(s) of the flow cell to process. Can be either a single lane, a range (e.g., 2-4) or '*' for all lanes in the flow cell. |
| Sample | The name of the sample. This name is the prefix to all the generated FASTQs, and corresponds to the --sample argument in all downstream 10x pipelines.Sample names must conform to the Illumina bcl2fastq naming requirements. Only letters, numbers, underscores and hyphens area allowed; no other symbols, including dots (".") are allowed. |
| Index | The 10x sample index that was used in library construction, e.g., SI-TT-A1 for a Dual-Indexed Multiome GEX library, or SI-NA-A1 for a Multiome ATAC library. |
To run mkfastq with a simple layout CSV, use the --csv
argument. Here's how to run mkfastq on the
tiny-bcl-atac sequencing run with the simple layout:
$ cellranger-arc mkfastq --id=tiny-bcl-atac \ --run=/path/to/cellranger-arc-tiny-bcl-atac-1.0.0 \ --csv=/path/to/cellranger-arc-tiny-bcl-atac-simple-1.0.0.csv cellranger-arc mkfastq (1.0.1) Copyright (c) 2020 10x Genomics, Inc. All rights reserved. ------------------------------------------------------------------------------- Martian Runtime - v4.0.1 Running preflight checks (please wait)... yyyy-mm-dd hh:mm:ss [runtime] (ready) ID.tiny-bcl-atac.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET yyyy-mm-dd hh:mm:ss [runtime] (split_complete) ID.tiny-bcl-atac.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET yyyy-mm-dd hh:mm:ss [runtime] (run:local) ID.tiny-bcl-atac.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET.fork0.chnk0.main yyyy-mm-dd hh:mm:ss [runtime] (chunks_complete) ID.tiny-bcl-atac.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET ...
Running mkfastq with an Illumina Experiment Manager Sample Sheet
The cellranger-arc mkfastq pipeline can also be run with a samplesheet in the Illumina Experiment Manager (IEM) format. If you didn't sequence with sample indices, you'll need to use this format. An IEM sample sheet consists of a number of fields specific to running on Illumina platforms, and then a [Data] section. That section is where you put your sample, lane and index information.
Here's an example of what the [Data] would look like for a dual-indexed Multiome GEX flow cell:
[Data] Lane,Sample_ID,Sample_Name,Sample_Plate,Sample_Well,I7_Index_ID,index,I5_Index_ID,index2,Sample_Project,Description 1,s1,test_sample,,,SI-TT-A1,SI-TT-A1,SI-TT-A1,SI-TT-A1,p1,
Here, SI-TT-A1 refers to a 10x dual-indexed library sample index. In this example, only reads from lane 1 will be used. To demultiplex the given sample index across all lanes, omit the lanes column entirely.
Here's an example of a Multiome ATAC flow cell that is single indexed:
[Data] Sample_ID,Sample_Name,Sample_Plate,Sample_Well,I7_Index_ID,index,Sample_Project,Description s1,test_sample_miseq,,,SI-NA-A1,SI-NA-A1,p1,
Here, SI-NA-A1 refers to a 10x single-indexed sample index, a set of four oligo sequences. cellranger-arc mkfastq also supports listing oligo sequences explicitly.
Sample names must conform to the Illumina bcl2fastq naming
requirements. Specifcally only letters, numbers, underscores and hyphens area
allowed. No other symbols, including dots (".") are allowed.
Also note that while an authentic IEM sample sheet will contain other sections above the [Data] section, these are optional for demultiplexing. For demultiplexing an existing run with cellranger-arc mkfastq, only the [Data] section is required.
Next, run the cellranger-arc mkfastq pipeline, using the --samplesheet argument:
$ cellranger-arc mkfastq --id=tiny-bcl-atac \ --run=/path/to/tiny-bcl-atac \ --samplesheet=cellranger-arc-tiny-bcl-atac-samplesheet-1.0.0.csv cellranger-arc mkfastq (1.0.1) Copyright (c) 2020 10x Genomics, Inc. All rights reserved. ------------------------------------------------------------------------------- Martian Runtime - v4.0.1 Running preflight checks (please wait)... yyyy-mm-dd hh:mm:ss [runtime] (ready) ID.tiny-bcl-atac.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET yyyy-mm-dd hh:mm:ss [runtime] (split_complete) ID.tiny-bcl-atac.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET yyyy-mm-dd hh:mm:ss [runtime] (run:local) ID.tiny-bcl-atac.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET.fork0.chnk0.main yyyy-mm-dd hh:mm:ss [runtime] (chunks_complete) ID.tiny-bcl-atac.MAKE_FASTQS_CS.MAKE_FASTQS.PREPARE_SAMPLESHEET ...
If you encounter any preflight errors, refer to the Troubleshooting page.
Checking FASTQ Output
Once the cellranger-arc mkfastq pipeline has successfully
completed, the output can be found in a new folder named with the value you
provided to cellranger-arc mkfastq in the --id option
(if not specified, defaults to the name of the flow cell):
$ cellranger-arc mkfastq --id=tiny-bcl-atac \ --run=/path/to/tiny-bcl-atac \ --samplesheet=cellranger-arc-tiny-bcl-atac-samplesheet-1.0.0.csv cellranger-arc mkfastq (1.0.1) Copyright (c) 2020 10x Genomics, Inc. All rights reserved. ------------------------------------------------------------------------------- Martian Runtime - v4.0.1 ... Pipestance completed successfully! yyyy-mm-dd hh:mm:ss Shutting down. Saving pipestance info to "tiny-bcl-atac/tiny-bcl-atac.mri.tgz" $ ls -l drwxrwxr-x 4 jdoe jdoe 4096 Aug 29 15:29 tiny-bcl-atac
The key output files can be found in outs/fastq_path, and are
organized in the same manner as a conventional bcl2fastq run:
$ ls -l tiny-bcl-atac/outs/fastq_path/ total 31744 drwxrwxr-x 3 jdoe jdoe 24 Sep 7 22:49 p1 drwxrwxr-x 3 jdoe jdoe 26 Sep 7 22:48 Reports drwxrwxr-x 2 jdoe jdoe 193 Sep 7 22:48 Stats -rw-rw-r-- 1 jdoe jdoe 3806257 Sep 7 22:48 Undetermined_S0_L001_I1_001.fastq.gz -rw-rw-r-- 1 jdoe jdoe 967448 Sep 7 22:48 Undetermined_S0_L001_R1_001.fastq.gz -rw-rw-r-- 1 jdoe jdoe 5773976 Sep 7 22:48 Undetermined_S0_L001_R2_001.fastq.gz -rw-rw-r-- 1 jdoe jdoe 12635207 Sep 7 22:48 Undetermined_S0_L001_R3_001.fastq.gz $ tree tiny-bcl-atac/outs/fastq_path/tiny-bcl-atac/ tiny-bcl-atac/outs/fastq_path/p1 └── s1 ├── test_sample_miseq_S1_L001_I1_001.fastq.gz ├── test_sample_miseq_S1_L001_R1_001.fastq.gz ├── test_sample_miseq_S1_L001_R2_001.fastq.gz └── test_sample_miseq_S1_L001_R3_001.fastq.gz
This example was produced with a sample sheet that included p1 as the Sample_Project, so the directory containing the sample folders is named p1. If a Sample_Project wasn't specified, or if a simple layout CSV file was used (which does not have a Sample_Project column), the directory containing the sample folders would be named according to the flow cell ID instead.
If you want to remove the Undetermined FASTQs from the output to
save space, you can run mkfastq with the
--delete-undetermined flag. To see all
cellranger-arc mkfastq options, run
cellranger-arc mkfastq --help.
Reading Quality Control Metrics
When the --qc flag is specified, the cellranger-arc
mkfastq pipeline writes both sequencing and 10x-specific quality control
metrics into a JSON file. The metrics are in the
outs/qc_summary.json file.
The use of --qc flag is not supported on NovaSeq flow cells.
|
The qc_summary.json file contains a number of useful metrics. The sample_qc key is a good place to start exploring your data.
"sample_qc": {
"Sample1": {
"5": {
"barcode_exact_match_ratio": 0.9336158258904611,
"barcode_q30_base_ratio": 0.9611993091728814,
"bc_on_whitelist": 0.9447542078230667,
"mean_barcode_qscore": 37.770630795934,
"number_reads": 2748155,
"read1_q30_base_ratio": 0.8947676653366835,
"read2_q30_base_ratio": 0.7771883245304577
},
"all": {
"barcode_exact_match_ratio": 0.9336158258904611,
"barcode_q30_base_ratio": 0.9611993091728814,
"bc_on_whitelist": 0.9447542078230667,
"mean_barcode_qscore": 37.770630795934,
"number_reads": 2748155,
"read1_q30_base_ratio": 0.8947676653366835,
"read2_q30_base_ratio": 0.7771883245304577
}
}
}
The sample_qc metric is a series of key value pairs for each sample in the sample sheet, and one metrics structure per lane per sample, plus an 'all' structure in case a sample spans multiple lanes.
The metrics are as follows:
| Key | Meaning |
barcode_exact_match_ratio | The percentage of barcode sequences that exactly match a whitelisted 10x barcode. |
barcode_q30_base_ratio | The percentage of barcode bases at or above Q30. |
bc_on_whitelist | The percentage of barcode sequences that match a 10x barcode on the whitelist, post error-correction. Corresponds to the "Valid Barcodes" value in cellranger-arc output metrics. |
mean_barcode_qscore | Mean quality score of barcode bases. |
number_reads | Reads per lane matching the sample's sample index (or overall in 'all'). |
read1_q30_base_ratio | The percentage of R1 bases at or above Q30. |
read2_q30_base_ratio | The percentage of R2 bases at or above Q30. |
By looking at this output, you can diagnose low barcode mapping rates and read
quality before running a cellranger-arc pipeline.
Additional metrics in outs/qc_summary.json include per-cycle
quality metrics, yield, cluster density and percent passing filter, and both
cellranger-arc and bcl2fastq version
information.
Troubleshooting
If you encounter a crash while running cellranger-arc mkfastq, upload the tarball (with the extension .mri.tgz) in your output directory:
cellranger-arc upload youremail@institution.edu jobid.mri.tgz
where jobid is what you input into the --id option of mkfastq (if not specified, defaults to the ID of the flow cell). This tarball contains numerous diagnostic logs that we can use for debugging.
You will receive an automated email from 10x Genomics. If not, email support@10xgenomics.com. For the fastest service, respond with the following:
- The exact command line you used.
- The sample sheet that you used.
- The RunInfo.xml and runParameters.xml files from your BCL directory.
- The kind of libraries you are demultiplexing (including chemistry).
- 2.0 (latest)
- Cell Ranger ARC v1.0