10x Genomics
Chromium Single Cell Multiome ATAC + Gene Exp.
Cell Ranger ARC1.0, printed on 07/28/2025
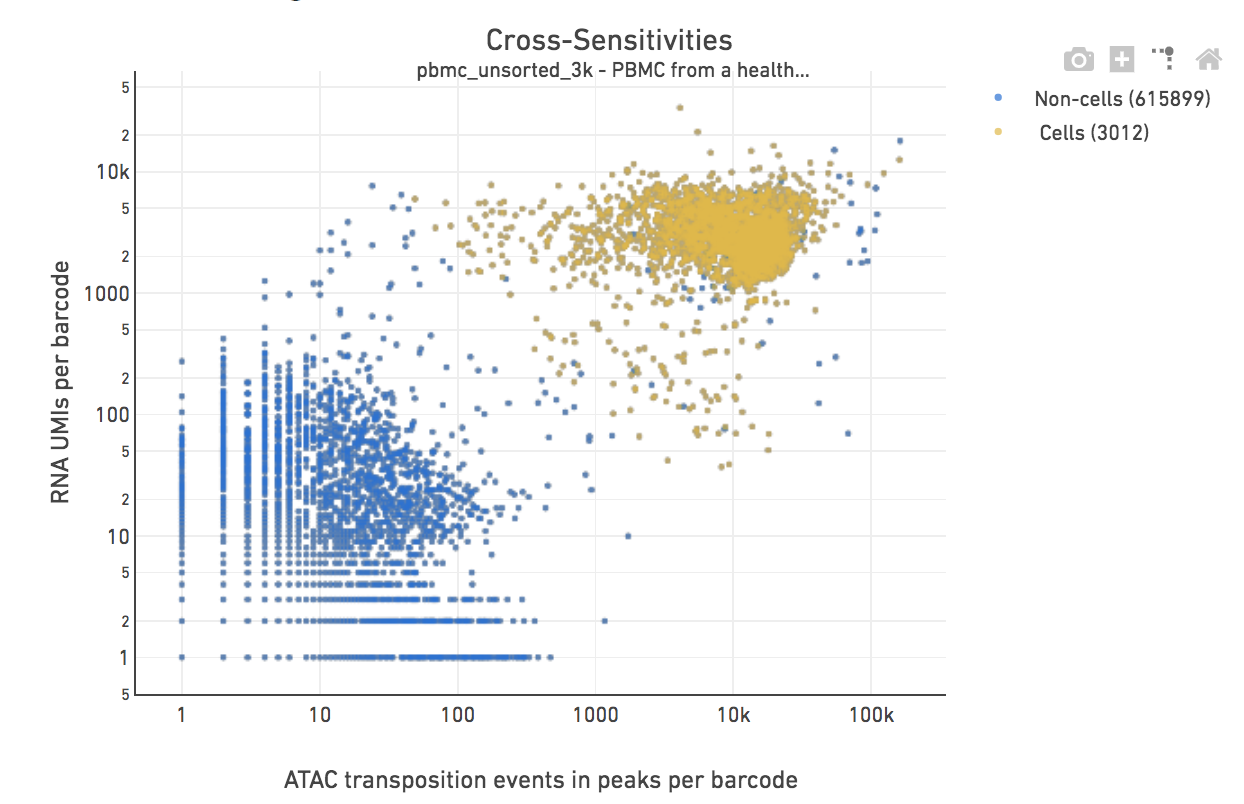
Calling Cell Barcodes
This step associates a subset of barcodes observed in the library to the nuclei loaded from the sample. Identification of these cell barcodes enables data analysis at single cell resolution. For each barcode, we have an associated count of transposition events in peaks (from the chromatin accessibility library), as well as gene expression UMIs (from the gene expression library). The cell-calling algorithm uses this paired information to identify the cell population from the non-cell background.
Prior to performing cell calling, barcodes must pass three filtering criteria.
First filter - ATAC "low targeting" barcodes: Identifies "low-targeting" barcodes in which the fraction of fragments overlapping called peaks is lower than the fraction of genome in peaks (for this calculation, peaks are padded by 2,000 bp on both sides to account for fragment length). These barcodes typically have their cut sites randomly distributed over the genome, are not targeted to be enriched near functional regions, and do not exhibit the typical ATAC-seq nucleosome phasing signal.
Second filter - gel bead doublets: Identifies putative gel bead doublets where a partition contains one cell and two barcoded gel beads. These cells then manifest as two barcodes of the same cell type in the dataset. The rate of such doublets is low and the presence of these few extra barcodes doesn't significantly affect secondary analysis such as clustering or differential analysis, although it can potentially inflate abundance measurements of very rare cell types. The chromatin accessibility library is used to identify a minor-major pair of barcodes (B1, B2) that are part of a putative gel bead doublet by observing if the pair of barcodes shares more genomically adjoining "linked" fragments (fragments sharing a transposition event) with each other (B1-B2) as opposed to themselves (B1-B1 or B2-B2). The minor barcode is identified as the one with fewer fragments.
Third filter - greater than 1 count observed in each library: Identifies barcodes that have been observed in both chromatin accessibility and gene expression libraries. The minimum threshold is set at >1 count in each library.
The first two filters rely solely on data from the chromatin accessibility library, and are often referred to as ATAC exclusions. A barcode that fails any of the above three filtering criteria is masked from the total set of barcodes prior to cell calling.
We then perform cell calling on the remainder barcodes. The algorithm has four key steps:
- De-duplication: The cell-calling algorithm places each barcode into a 2-D space defined by their individual ATAC and GEX counts. Barcodes with identical coordinates i.e. ATAC and GEX counts are collapsed into a single measurement to generate a more uniform density of points across the 2-D space. This de-duplication step de-emphasizes over-represented low-count barcodes and allows suppression of noise without using thresholds or making assumptions about the count distribution profiles.
- Ordmag-derived initial grouping: The de-duplicated count data is partitioned using thresholds derived from “ordmag” into cell and non-cell populations. Ordmag is a published algorithm that finds a threshold that is 10-fold less than the maximum value after removing outliers. A threshold is defined independently for each dimension and barcodes above both ATAC and GEX thresholds are labeled as cells, with the remainder labeled as non-cells.
- K-means boundary refinement: Since “ordmag” is insufficient to capture the spread of paired ATAC and GEX data, we refine the boundaries of these initial set of cells using K-means. The K-means algorithm is initialized using centroids calculated from the ordmag-defined cell and non-cell groups. The K-means algorithm is implemented using this single initialization and set to K=2.
- Map classification to de-duplicated barcodes: The K-means assignment of the de-duplicated count data into cells and non-cells are used to classify the full set of non-excluded barcodes. Barcodes that were masked during the de-duplication step are assigned to the cell or non-cell group based on the K-means classification of their counterpart with identical ATAC and GEX counts.
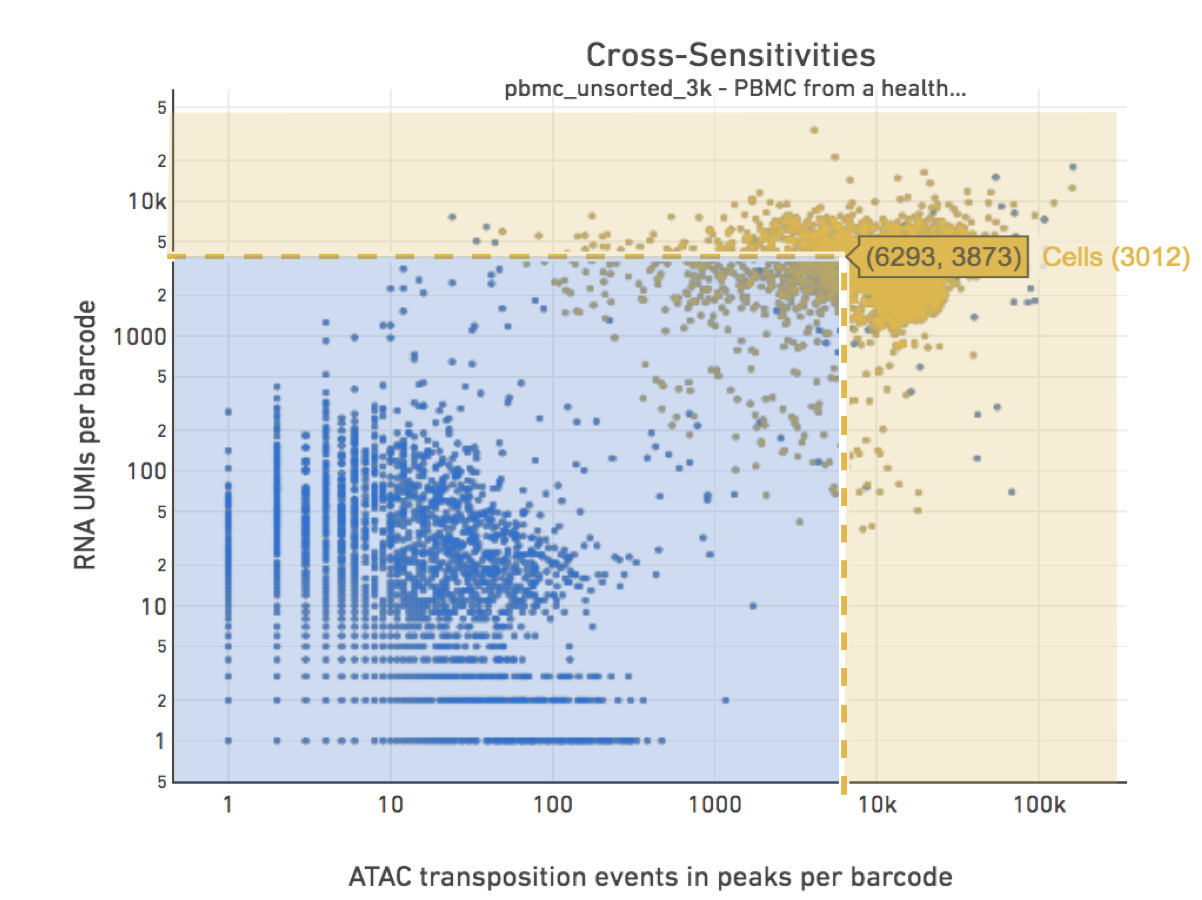
Cell calling is limited to produce < 20k cells as the assay is currently designed to support 500-10k cells. To override the cell-caller, two additional parameters need to be provided to cellranger-arc count: --min-atac-count=N and --min-gex-count=N, the former specifies the minimum number of transposition events in peaks for a cell barcode and the latter specifies the minimum number of UMI counts for a cell barcode. Any barcode that satisfies either threshold will be classified as a cell, unless it is excluded from the cell calling algorithm because it fails the above specified filtering criteria.
The figure below shows how Force Cells can be used to override the default pipeline Joint Cell Calling algorithm. With override parameters set to values specified by the drop-spike-lines: --min-atac-count=6293 and --min-gex-count=3873, barcodes in the shaded blue area will be classified as non-cell barcodes and those in the yellow shaded area as cells.
 |
 |
- 2.0 (latest)
- Cell Ranger ARC v1.0