10x Genomics
Chromium Single Cell CNV
Cell Ranger DNA1.0, printed on 07/26/2025
Preprocessing
Extract read barcode
The first 16 bases of read 1 contain the 10x barcode, which identifies the partition from which the DNA originates. The barcode by design can take one of 737,000 different sequences that comprise a whitelist. This feature allows us to perform error correction when the observed barcode does not match any barcode on the whitelist due to sequencing error.
Align and mark duplicates
After trimming the barcode sequence, we align all the trimmed read pairs to the reference genome using BWA-MEM with flag -M.
After alignment we mark duplicate read-pairs using the heuristic that when two read-pairs with the same barcode align to the same fragment on the reference genome they are duplicates of each other.
This mark is encoded in the PCR or optical duplicate flag of each alignment record in the BAM output.
Define cell barcodes
Each barcode labels a partition but not every partition contains a cell. A small fraction of reads are associated with empty partitions as a consequence of either the library creation process, sequencing or barcode error correction. To identify the partitions containing cells or cell barcodes we calculate the distribution of non-duplicate reads with mapping quality at least 30 per barcode.
The figure below shows the reads per barcode for all observed barcodes in a sample sorted such that the barcode with the most reads appears first. The steep drop in the reads per barcode is indicative of a good separation between cell barcodes and non-cell barcodes. The barcodes on the left of the drop are cells and those on the right are noise.
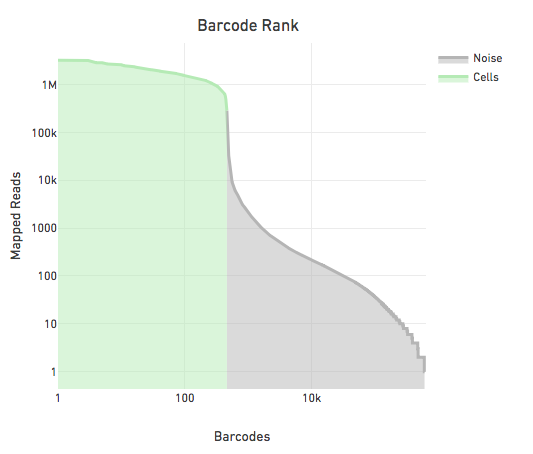
The large separation between cell barcodes and non cell barcodes allows for the following simple heuristic to identify cell barcodes:
- we order the barcodes by the number of reads in ascending order
- we select the barcodes that contain at least 1/10th as many reads as the last barcode. This gives us an initial set enriched for cell barcodes.
- we calculate the 99-th percentile of the reads per barcode distribution restricted to the set of barcodes identified in the previous step
- we define all barcodes with at least 1/10th as many reads as the 99-th percentile as cell barcodes
In rare situations this heuristic can fail to capture all the cells in the sample. Hypothetically, if 50% of the cell population were diploid and 50% of the cell population was hexaploid, then the 99-th percentile in the above heuristic would be based on the hexaploid cells alone. This could potentially exclude a few low coverage diploid cells. In such situations, the user can supply the argument --force-cells=N that forces the the top N barcodes, in terms of read counts, to be regarded as cell barcodes.
Compute coverage profile matrix
From the BAM file we compute the read-pair coverage over the genome for each cell barcode using only read-pairs that have mapping quality at least 30 and are not marked duplicates. The coverage is computed over 20 kb bins over the genome and is calculated as follows. When both reads in a pair map to the same chromosome and the size of the insert represented by the read-pair is less than 20 kb each read contributes 0.5 to the bin it maps to. When only one read in a pair is mapped the read contributes 1.0 to the bin it maps to. When the insert size is greater than 20 kb or when the two reads in a pair map to different chromosomes, each read contributes 1.0 to the bin it maps to. The coverage profile is represented as a set of matrices for each chromosome where the cells are the rows and the contiguous 20 kb bins on a chromosome are columns.
For a typical human sample sequenced to a depth of 750,000 read pairs per cell (1.5 million reads per cell), the average coverage per 20 kb bin is approximately in the range of 3-4 read pairs.
Compute mappability and GC content of the reference genome
We simulate paired-end perfect reads from the reference genome at 1X coverage with read length and insert size determined by the input library. We divide the genome into 20 kb bins and for each bin we calculate the fraction of simulated read-pairs that map back to the bin with mapping quality at least 30. This defines the mappability of each bin. Regions of the reference genome that are entirely composed of N bases would have a mappability of zero, while regions containing sequence that is unique to the genome will have a mappability of one. We also calculate the GC content per bin, i.e., the fraction of bases in each 20 kb bin that are G or C.
The coverage profile along with the mappability and GC are the sole inputs to the copy number calling algorithm described in the next section.
- 1.1 (latest)
- Cell Ranger DNA v1.0