10x Genomics
Visium Spatial Gene Expression
Space Ranger1.2, printed on 07/09/2025
Image Outputs
The pipeline output contains a subdirectory named spatial which stores imaging related files. The files include the
following:
-
tissue_hires_image.pngandtissue_lowres_image.png: These files are downsampled versions of the original, full-resolution brightfield image provided by the user. Downsampling is accomplished by box filtering, which averages RGB values in patches of pixels in the full-resolution image to obtain an RGB value of one pixel in the downsampled image. Thetissue_hires_image.pngimage has 2,000 pixels in its largest dimension, and thetissue_lowres_image.pnghas 600 pixels. -
aligned_fiducials.jpg: This image has the dimensions oftissue_hires_image.png. Fiducial spots found by the fiducial alignment algorithm are highlighted in red. This file is useful to verify that fiducial alignment was successful.

-
scalefactors_json.json: This file contains the following fields:tissue_hires_scalef: A scaling factor that converts pixel positions in the original, full-resolution image to pixel positions intissue_hires_image.png.tissue_lowres_scalef: A scaling factor that converts pixel positions in the original, full-resolution image to pixel positions intissue_lowres_image.png.fiducial_diameter_fullres: The number of pixels that span the diameter of a fiducial spot in the original, full-resolution image.spot_diameter_fullres: The number of pixels that span the diameter of a tissue spot in the original, full-resolution image.
$ cd /home/jdoe/runs/sample345/outs/spatial $ cat scalefactors_json.json {"spot_diameter_fullres": 89.45248682925602, "tissue_hires_scalef": 0.17699115, "fiducial_diameter_fullres": 144.5001710318751, "tissue_lowres_scalef": 0.053097345}
-
detected_tissue_image.jpg: This image has the dimensions of thetissue_hires_image.pngand shows the following:- Aligned fiducial spots as blue, hollow circles.
- A blue bounding box. The interior designates the portion of the image where tissue is discriminated from background.
- Spots found under tissue are red.
- Spots outside of tissue are gray.
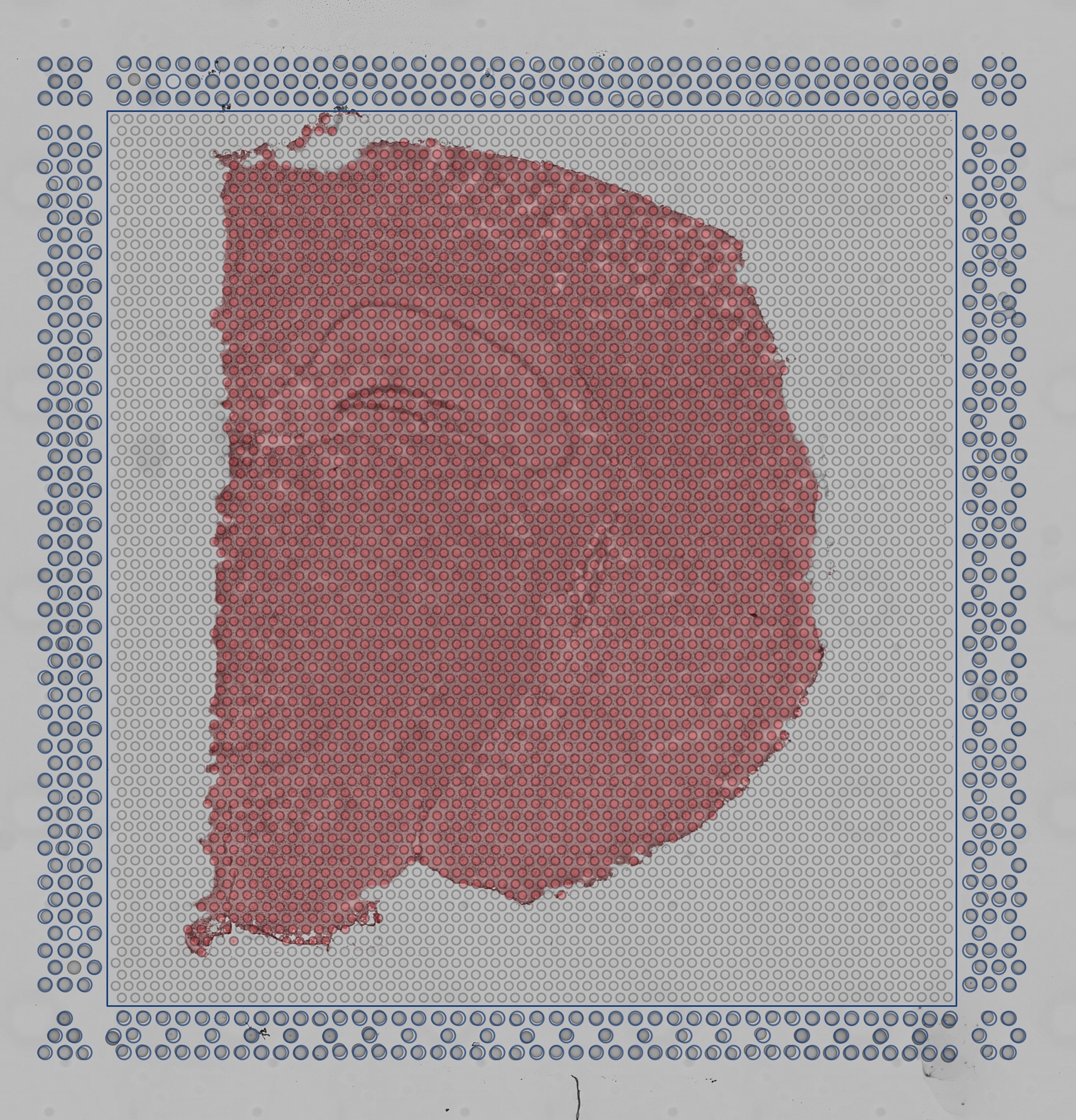
-
tissue_positions_list.csv: This text file contains a table with rows that correspond to spots. It has 4,992 rows, which is the number of spots in the spatial array. Columns, whose names are not specified in the file, correspond to the following fields:barcode: The sequence of the barcode associated to the spot.in_tissue: Binary, indicating if the spot falls inside (1) or outside (0) of tissue.array_row: The row coordinate of the spot in the array from 0 to 77. The array has 78 rows.array_col: The column coordinate of the spot in the array. In order to express the orange crate arrangement of the spots, this column index uses even numbers from 0 to 126 for even rows, and odd numbers from 1 to 127 for odd rows. Notice then that each row (even or odd) has 64 spots.pxl_row_in_fullres: The row pixel coordinate of the center of the spot in the full resolution image.pxl_col_in_fullres: The column pixel coordinate of the center of the spot in the full resolution image.
$ cd /home/jdoe/runs/sample345/outs/spatial $ head -2 tissue_positions_list.csv ACGCCTGACACGCGCT-1,0,0,0,910,1261 TACCGATCCAACACTT-1,0,1,1,1030,1329
- 1.0
- 1.1
- 1.3
- 2.0 (latest)
- Space Ranger v1.2