10x Genomics
Chromium Single Cell Immune Profiling
Cell Ranger2.0, printed on 07/09/2025
What is Cell Ranger for V(D)J?
Cell Ranger for V(D)J is a set of analysis pipelines that process Chromium single cell 5’ RNA-seq output to assemble, quantify, and annotate paired V(D)J transcript sequences.
Pipelines
Cell Ranger for V(D)J includes two main pipelines:
-
cellranger mkfastq wraps Illumina's bcl2fastq to correctly demultiplex Chromium-prepared sequencing samples and to convert barcode and read data to FASTQ files.
-
cellranger vdj takes FASTQ files from cellranger mkfastq and performs V(D)J sequence assembly and paired clonotype calling. It uses the Chromium cellular barcodes and UMIs to assemble V(D)J transcripts cell-by-cell. cellranger can take input from multiple sequencing runs on the same library and/or from multiple libraries from the same sample.
Output is delivered in standard BAM, CSV, FASTA, FASTQ, JSON and HTML formats that are augmented with cellular and clonotype-specific information.
Terminology
Throughout the documentation, you will see references to samples, libraries and sequencing runs. We define these as follows:
- Sample: A cell suspension extracted from a single biological source (blood, tissue, etc).
- Library: A 10x-barcoded sequencing library prepared from a single sample, corresponding to a single chip channel of a 10x Chromium Controller run.
- Sequencing Run / Flowcell: A flowcell containing data from one sequencing instrument run. The sequencing data can be further addressed by lane and by one or more sample indices.
The relationship between these terms can be complex:
- A single sample may be prepared into multiple libraries; this is commonly done to achieve higher cell counts without overloading a single 10x Chromium Controller run.
- A single library may be sequenced across multiple flowcells, and then combined as if all the reads came from one sequencing run.
- A single flowcell may contain multiple libraries, separated using different lanes and / or sample indices.
- The definition of "library" makes no distinction between two channels of the same physical 10x chip, versus the same channel of two different chips.
Workflows
The Cell Ranger workflow always starts with running cellranger mkfastq on each flowcell directory, as described in Generating FASTQs. The subsequent steps vary depending on how many samples, libraries and flowcells you have. We will describe them in order of increasing complexity:
Single Sample, Library, and Flowcell
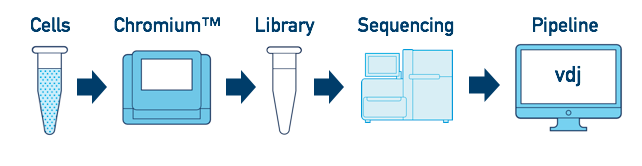
This is the most basic case. You have a single biological sample, which was prepared into a single library, and then sequenced on a single flowcell. Assuming the FASTQs have been generated with cellranger mkfastq, you just need to run cellranger vdj as described in V(D)J T Cell Analysis.
One Library, Multiple Flowcells
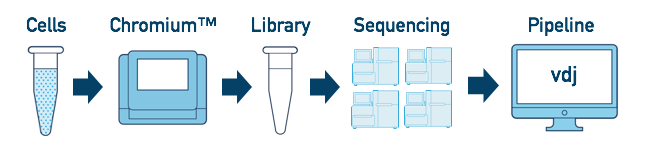
If you have a library which was sequenced across multiple flowcells, you can pool the reads from both sequencing runs. Follow the steps in Multi-Flowcell Samples to combine them in a single cellranger vdj run.
Multiple Libraries from the Same Biological Sample
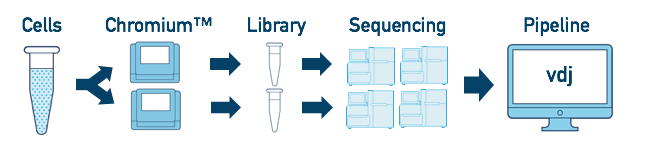
If you have a single sample which was run on multiple chip channels, producing multiple libraries, you can analyze all of the libraries at once. The workflow is similar to the "Multiple Flowcell" workflow above. Follow the steps in Multi-Flowcell Samples to combine them in a single cellranger vdj run.