10x Genomics
Single Cell Multiome ATAC + Gene Exp.
Cell Ranger ARC, printed on 07/16/2025
What's New in Loupe Browser 4.2
Loupe Browser 4.2 has been updated to allow users to easily analyze data
generated with the Single Cell Multiome ATAC + Gene Expression assay and
Cell Ranger ARC 1.0 pipeline, all while still allowing users to easily explore
single-cell gene expression, immunology, ATAC and spatial genomics data.
It is compatible with Cell Ranger ARC 1.0, and remains compatible with .cloupe
files generated by all versions of Space Ranger, Cell Ranger ATAC, and
Cell Ranger after v1.3.
Multiome ATAC + Gene Expression Support
Loupe Browser 4.2 allows users to directly analyze the relation between gene expression and chromatin accessibility from the same cell, using a variety of intuitive, interactive tools:
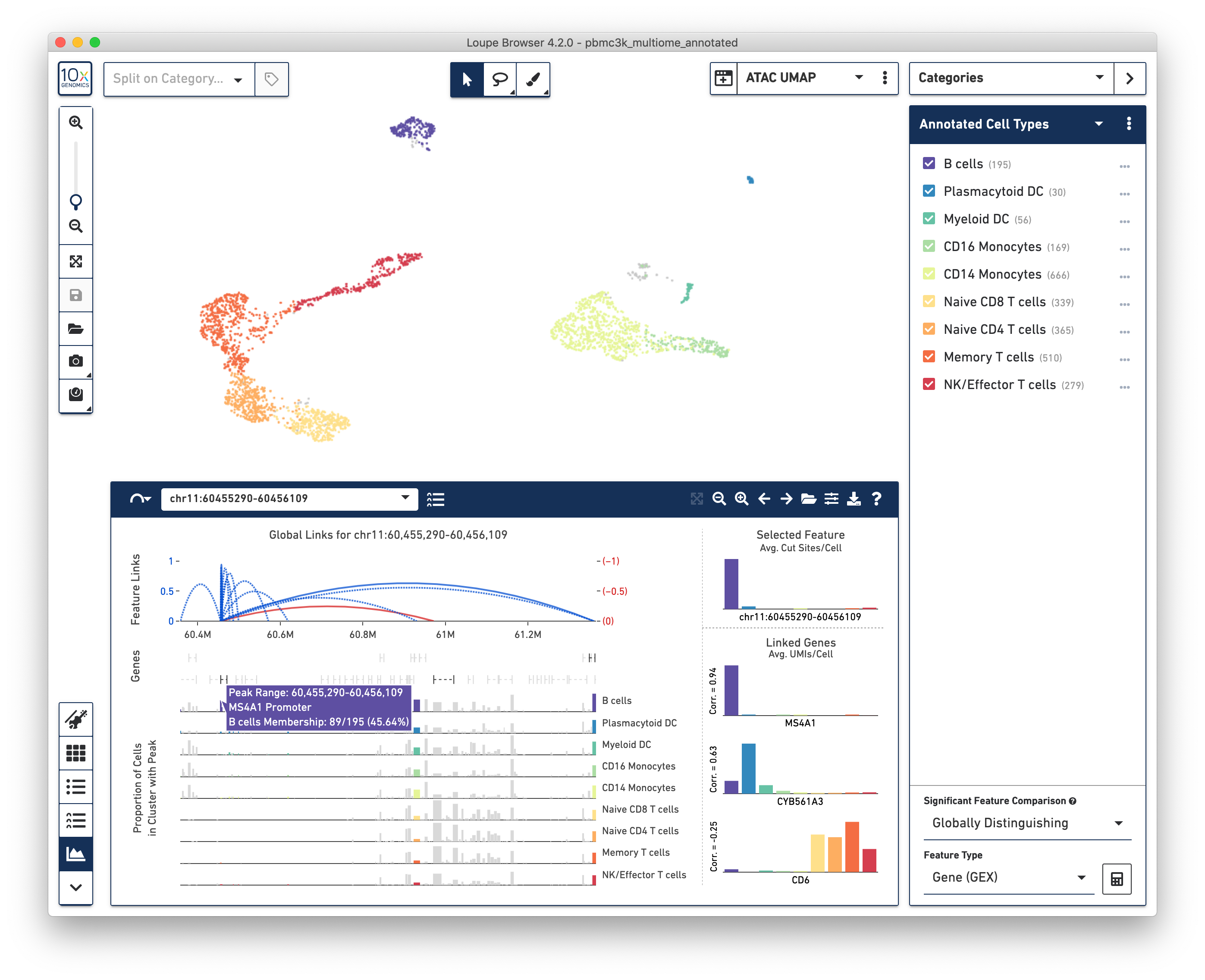
In particular, a new Feature Linkage Table and Feature Linkage View allows users to quickly discover transcriptional regions that may regulate gene expression, and intuit possible transcriptional networks. Users may also look at gene expression and ATAC-derived clusters, either within the same projection through split view, or side by side with linked windows.
To find out how to use Loupe Browser 4.2 to analyze true multiomic ATAC gene expression data, please consult the Loupe Browser documentation.
Improvements For All Applications
Split View
The Split View menu has been streamlined, with the ability to quickly select any clustering to split by, and easy toggling of cluster labels.