10x Genomics
Chromium Single Cell Multiome ATAC + Gene Exp.
Cell Ranger ARC2.0 (latest), printed on 08/08/2025
What is Cell Ranger ARC?
Cell Ranger ARC is a set of analysis pipelines that process Chromium Single Cell Multiome ATAC + Gene Expression sequencing data to generate a variety of analyses pertaining to gene expression (GEX), chromatin accessibility, and their linkage. Furthermore, since the ATAC and GEX measurements are on the very same cell, we are able to perform analyses that link chromatin accessibility and GEX.
-
cellranger-arc mkfastq demultiplexes raw base call (BCL) files generated by Illumina sequencers into FASTQ files. It is a wrapper around Illumina's bcl2fastq, with additional useful features that are specific to 10x Genomics libraries and a simplified sample sheet format. The same command can be used to demultiplex both ATAC and GEX flow cells.
-
cellranger-arc count takes FASTQ files from cellranger-arc mkfastq and performs alignment, filtering, barcode counting, peak calling, and counting of both ATAC and GEX molecules. Furthermore, it uses the Chromium cellular barcodes to generate feature-barcode matrices, perform dimensionality reduction, determine clusters, perform differential analysis on clusters, and identify linkages between peaks and genes. The count pipeline can take input from multiple sequencing runs on the same GEM well.
-
cellranger-arc aggr aggregates and analyzes the outputs from multiple runs of cellranger-arc count (such as from multiple samples from one experiment). Features include normalization of input runs to same median fragments per cell (sensitivity), detection of accessible chromatin peaks, count matrix generation for peaks and transcription factors for the aggregate data, dimensionality reduction, cell clustering, and cluster differential accessibility analysis.
-
cellranger-arc reanalyze takes the analysis files produced by cellranger-arc count or cellranger-arc aggr and reruns secondary analysis. Features include tunable parameter settings related to cell calling, dimensionality reduction, cell clustering, and cluster differential accessibility analysis.
These pipelines combine Chromium-specific algorithms with the widely
used aligners STAR and
BWA. Output is delivered in standard BAM,
MEX, CSV, HDF5, and HTML formats that are augmented with cellular information and
a .cloupe file for use with Loupe Browser.
Ways to run Cell Ranger ARC
- Run Cell Ranger ARC on 10x Genomics Cloud Analysis
Skip Cell Ranger ARC download and installation and get started with 10x Genomics Cloud Analysis, our recommended method for running Cell Ranger ARC pipelines for most new customers. Use your web browser to easily generate Cell Ranger ARC outputs from your FASTQ files and aggregate outputs from multiple runs, free for every 10x Genomics sample. Currently available only in the United States and Canada. Sign up for a free account or view tutorials and learn more.
- Install and run Cell Ranger ARC on your own computing infrastructure
Learn how to install and run Cell Ranger ARC.
Workflows
The Cell Ranger ARC workflow starts with demultiplexing the BCL files for each flow cell directory for all relevant ATAC and GEX sequencing runs. 10x Genomics recommends using cellranger-arc mkfastq as described in Generating FASTQs. If you are beginning with FASTQ files that have already been demultiplexed with bcl2fastq or BCL convert directly, or from a public source such as SRA, skip cellranger-arc mkfastq and begin with cellranger-arc count. The Specifying Input FASTQs page has specific guidelines on which arguments to use for your scenario.
The exact steps of the workflow vary depending on the number of samples, GEM wells, and flow cells used. This section describes the simplest possible workflows.
One sample, one GEM well, one ATAC + one GEX flow cell
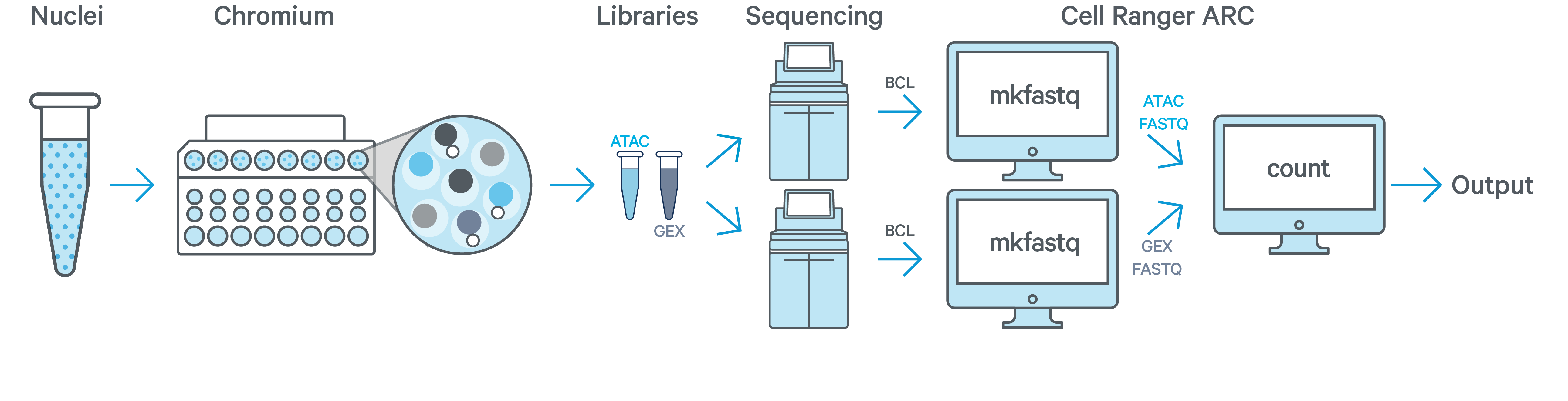
In this example you have one sample that is processed through one GEM well (a set of partitioned cells from a single 10x Chromium™ Chip channel) and results in one Multiome ATAC library and one Multiome GEX library. Each library is sequenced separately on one flow cell. In this case you would generate FASTQs separately for ATAC and GEX by running cellranger-arc mkfastq on the respective flow cells and run cellranger-arc count as described in Single-Sample Analysis.
One sample, one GEM well, multiple ATAC + multiple GEX flow cells
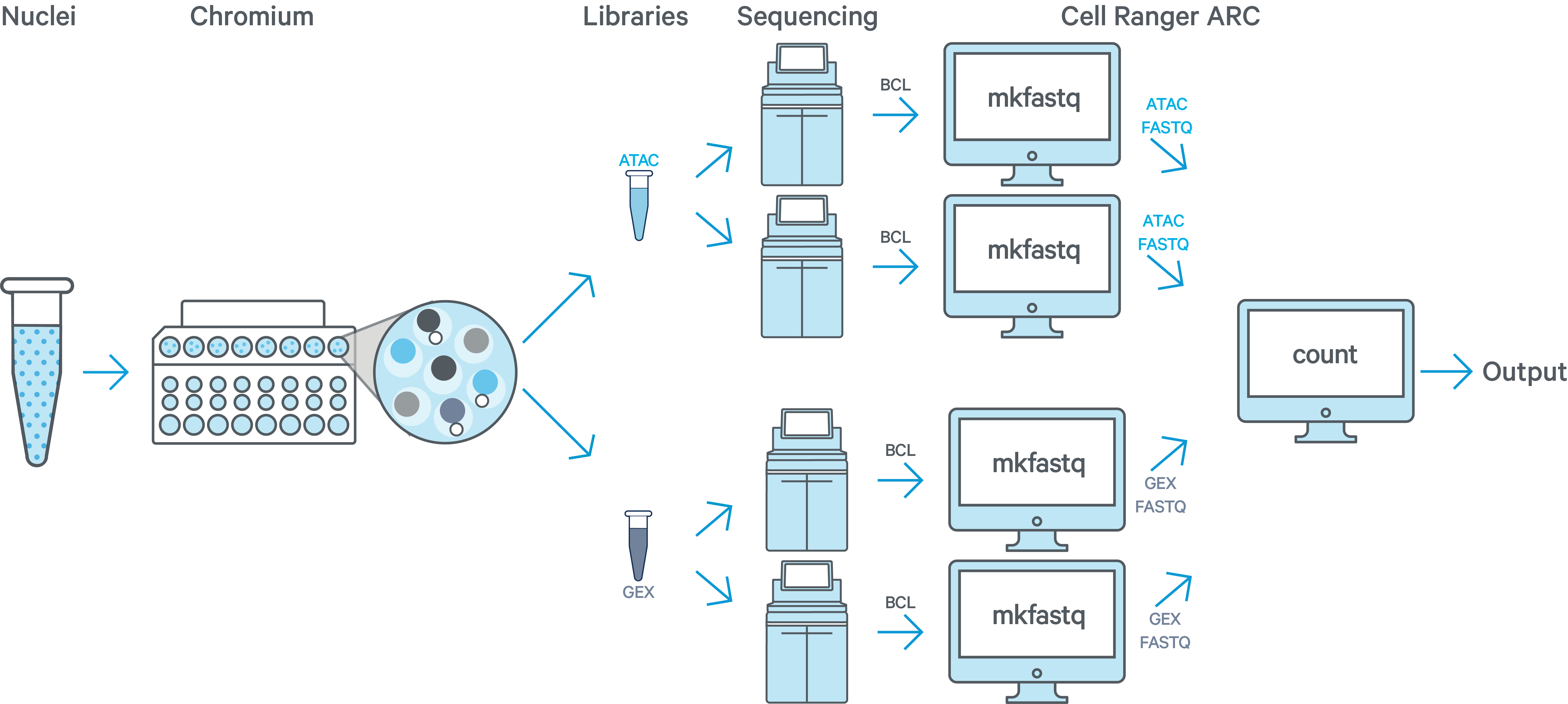
In this example you have one sample that is processed through one GEM well resulting in one ATAC library and one GEX library. The ATAC and GEX libraries are sequenced on two flow cells each. As an example, this may be done to increase sequencing depth, when the first sequencing run did not produce enough raw read pairs per cell. Here we would run cellranger-arc mkfastq a total of four times: once for each of the two ATAC flow cells and once for each of the two GEX flow cells. All of the reads can be combined in a single instance of the cellranger-arc count pipeline. This process is described in Specifying Input Fastqs.
Multiple samples, multiple GEM wells, one ATAC + one GEX flow cell
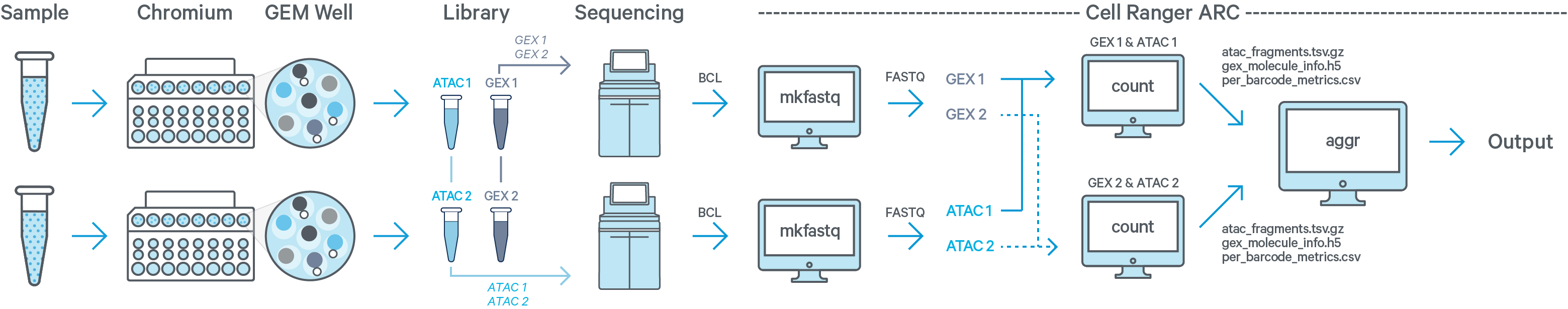
In this example you have two samples that were processed through two GEM wells resulting in one ATAC library and one GEX library per GEM well. The two ATAC libraries are sequenced together on a flow cell, and the two GEX libraries are sequenced together on a different flow cell. Then run cellranger-arc mkfastq twice: once for the ATAC flow cell and once for the GEX flow cell. The resulting ATAC + GEX FASTQ files from sample 1 are input into one instance of the cellranger-arc count pipeline. Similarly, ATAC + GEX FASTQs from sample 2 are processed together in a second instance of cellranger-arc count. This process is described in Specifying Input Fastqs.
- 1.0
- Cell Ranger ARC v2.0 (latest)