10x Genomics
Chromium Genome & Exome
Long Ranger2.2 (latest), printed on 07/08/2025
Using GATK with Long Ranger
| Analysis software for 10x Genomics linked read products is no longer supported. Raw data processing pipelines and visualization tools are available for download and can be used for analyzing legacy data from 10x Genomics kits in accordance with our end user licensing agreement without support. |
We recommend running longranger with GATK for more accurate calling of SNPs and indels. You must already have a working version of GATK installed on your system. Support is also provided for using a bundled build of Freebayes. Long Ranger uses GATK's HaplotypeCaller mode. See the GATK installation instructions
Performance Comparison
The GATK mode of Long Ranger yields substantially more accurate indel calls. Below are ROC curves reported by hap.py for 30x WGS Chromium data on NA12878, against the Genome In a Bottle v3.2.2 reference variants. GATK improves indel sensitivity by ~5 percentage points, while improving specificty.
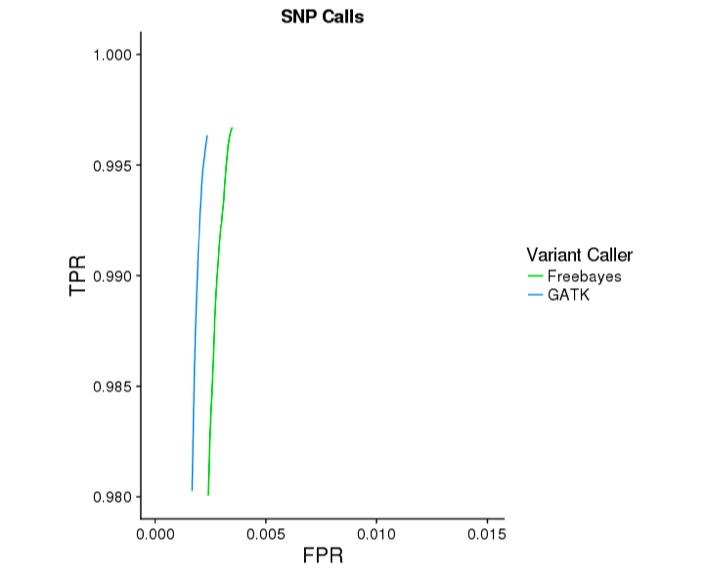
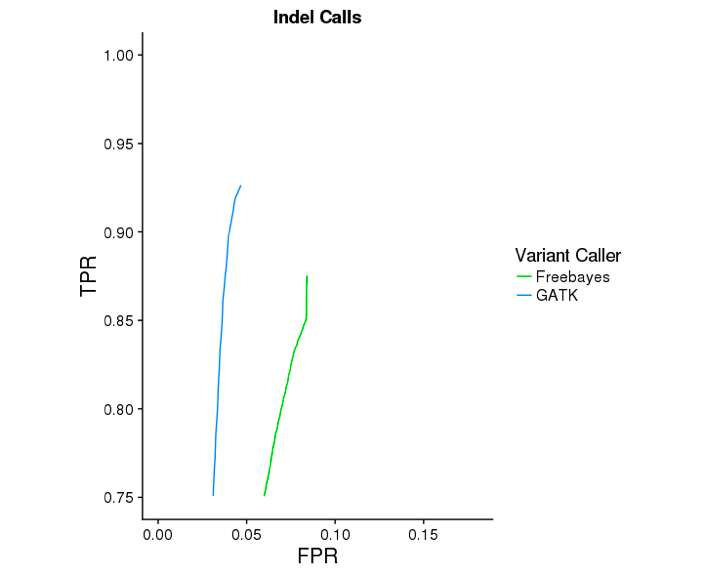
Version Compatibility
| Long Ranger Version | GATK Version Supported |
|---|---|
| 2.2 and later | 3.3-4.0, excluding 3.6 |
| 2.1 and earlier | 3.3-3.8, excluding 3.6 |
| Changes to GATK occurring after version 4.0.3 have introduced an incompatibility with Long Ranger. We will release an update to Long Ranger to account for this, but in the meantime the maximum version level for GATK that will work is 4.0.3. You can download GATK 4.0.3 from here. |
Configuring GATK
To use GATK, please note the following:
- You must already have a working version of GATK installed on your system; the Long Ranger software does not include the GATK distribution.
- You must use a version of GATK that is compatible with your version of Long Ranger; see the Version Compatibility table, above. Note that GATK version 3.6 has a crashing bug in the 'single-haplotype' mode, which Long Ranger uses, and so is not compatible..
- If you are using a custom reference,
longranger mkrefwill instruct you on how to create agenome.dictfile. You will need this file to use GATK.
You can instruct longranger to use GATK as its variant caller by passing the --vcmode=gatk:/path/to/GenomeAnalysisTK.jar parameter. For example, if your GenomeAnalysisTK.jar jarfile is located in /usr/local/gatk/GenomeAnalysisTK.jar,
$ longranger wgs --id=sample345 \ --sex=female \ --fastqs=/home/jdoe/runs/HAWT7ADXX/outs/fastq_path \ --indices=SI-GA-A1 \ --reference=/opt/refdata-hg19-2.1.0 \ --targets=/home/jdoe/runs/agilent_exome.bed \ --vcmode=gatk:/usr/local/gatk/GenomeAnalysisTK.jar \ --uiport=3600
Aside from the stage where SNPs and indels are called, Long Ranger will behave the same whether using GATK or Freebayes.
| The path to the GATK JAR must be an absolute path (e.g., /home/jdoe/bin/GenomeAnalysisTK.jar, not just bin/GenomeAnalysisTK.jar). |